
Contact Information
Research Interests
Research Topics
Regulation of Gene Expression, RNA Biology
Disease Research Interests
Aging Related Diseases, Cancer
Research Description
Long-noncoding RNA; Gene regulation; Nuclear domain & structure, breast cancer
The human genome encodes thousands of long noncoding RNA (lncRNA) genes. Although genetic studies have unraveled the function of a subset of these lncRNAs in key cellular processes, we still lack mechanistic insights into the role of most lncRNAs. Based on studies on a handful of them, we learned that lncRNAs regulate vital cellular processes by functioning as ‘scaffolds’ or ‘guides’ or ‘sponges’ to modulate protein-protein or protein-DNA or protein-RNA interactions, or as ‘enhancer RNAs’ to regulate gene expression. Nuclear lncRNAs also act as nucleation sites for the formation and/or maintenance of sub-nuclear domains (NDs). Further, lncRNAs show deregulated expression in tumors and their genes are often amplified or deleted in various cancers, undermining the importance of understanding the molecular and cellular roles of lncRNAs under physiological as well as pathological conditions.
The long-term goal of my laboratory is to understand how lncRNAs localized within nuclear domains control gene expression, particularly during cancer progression. By utilizing cellular and molecular biological approaches, we have unraveled the function of several lncRNAs in mammalian cells and their involvement in cancer progression. Our current research continues to dissect the role of lncRNAs in gene regulatory mechanisms. Specifically, we are studying: i) how lncRNAs enriched in discrete nuclear domains (NDs) regulate gene expression through their modulation of ND and associated chromatin structure, ii) how cell cycle-regulated lncRNAs may regulate cell proliferation and cancer progression, and iii) how cancer-deregulated lncRNAs contribute tumor progression and cancer metastasis.
i. Characterization of nuclear domain-enriched RNA-mediated gene regulatory mechanisms
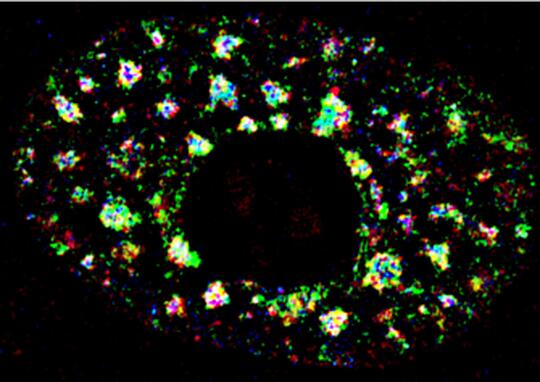
In order to understand the role of lncRNAs in various nuclear processes, we focused our efforts in understanding the role of lncRNAs enriched in nuclear domains such as nuclear speckles, paraspeckles, and nucleolus. We demonstrated that nuclear speckle-localized RNA and proteins display multilayer organization, and MALAT1regulates the speckle distribution and splicing function of SR family of splicing factors (SRSFs) (Fig. 1) (Fei et al., J. Cell Sci. 2017; Tripathi et al., Mol Cell 2010; MBoC, 2013). In the case of paraspeckles, we observed that Adenosine-to-Inosine (A-I) editing of RNA is not essential for RNA to be localized in paraspeckles (Anantharaman et al., Sci Rep 2016). Furthermore, we observed that A-to-I editing regulates the stability of a subset of mRNA by influencing the interaction between the RNA and RNA-destabilization factors (Anantharaman et al., NAR 2017; Anantharaman et al., FEBs Lett. 2017).
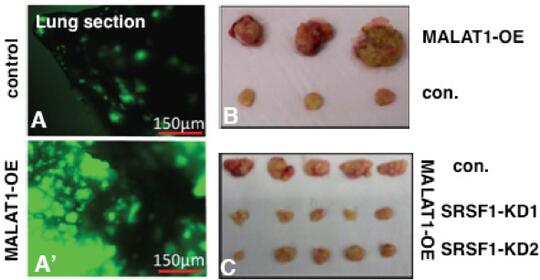
MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript1) serves as a model lncRNA for us to establish the connection between a ND-enriched lncRNA and cancer. MALAT1 is a long (~7kb), abundant, highly conserved nuclear lncRNA that is enriched in nuclear speckle domains. Depletion of MALAT1 reduced tumorigenic and metastatic properties of cancer cells, whereas its overexpression dramatically enhanced tumor lung colonization in mouse xenograft models (Jadaliha et al., Oncotarget 2016; Malakar et al., Cancer Res. 2017) (Fig. 2). Although MALAT1’s involvement in cancer progression and metastasis is undisputed, insights into the molecular mechanism underlying MALAT1’s physiological and pathological roles remain largely unknown. We were the first to demonstrate the involvement of MALAT1in AS (alternative splicing) of pre-mRNAs and splicing factor distribution in speckles (Tripathi et al., Mol Cell 2010; Bernard et al., EMBO J 2010; Tripathi et al., MBoC 2012; PloS Genet. 2013). However, the molecular basis of how MALAT1 regulates alternative splicing (AS) remains unknown. The goal of my laboratory is to fill this major gap and to establish the molecular mechanisms underlying MALAT1-regulated alternative splicing during breast cancer progression. We recently demonstrated the involvement of MALAT1 in organizing splicing factors such as SRSFs in speckles. By performing super-resolution imaging analyses, we observed that MALAT1 and several other pre-mRNA splicing factors display non-random localization within speckles, and such localization of factors seems to influence speckle function (Fei et al., 2017). We will determine whether phase separation of speckle components play crucial role in the multilayer organization of factors within speckles. Mechanistically, we will test the model that MALAT1 regulates alternative splicing by modulating speckle localization of splicing factors and molecular interactions between splicing factors and other speckle-resident proteins. In general, characterization of speckle-associated proteins and the transcriptome would unravel the principles that define the contribution of nuclear speckles in genome organization and RNA maturation under physiological and pathological conditions.
ii. Characterization of cell cycle-regulated lncRNAs involved in cell proliferation
The cell cycle is a vital cellular process, subject to stringent control and frequently perturbed by cancer-inducing mutations. The activity of genes controlling cell proliferation is modulated through the cell cycle dependent oscillation of their transcription and/or post-translational modifications. Recent studies indicate that lncRNAs control cell proliferation and cell survival, by regulating the expression of cell cycle-regulated protein-coding genes, underscoring the importance of lncRNAs in regulating cell proliferation. By performing transcriptome-wide RNA-seq screens in cells synchronized at different stages of the cell cycle we identified several hundreds of lncRNAs that display cell cycle stage-specific expression. Furthermore, we observed that several of the lncRNAs contain microRNAs within them; however, we have begun to elucidate a microRNA-independent role of these lncRNAs in cell cycle progression (Sun et al., NAR, 2018). Additionally, we found that some of the candidate lncRNA genes are located in close proximity to protein-coding genes. These lncRNAs regulate transcription and RNA processing of their protein-coding gene partners, and control cell proliferation by governing vital pathways such as HIPPO signaling (Hao et al., 2020). We plan to functionally characterize several of these cell cycle-regulated lncRNAs to determine their role in specific cell cycle stages and/or signaling pathways that control cell cycle checkpoints.
iii. Characterization of lncRNAs deregulated during breast cancer (BC) progression
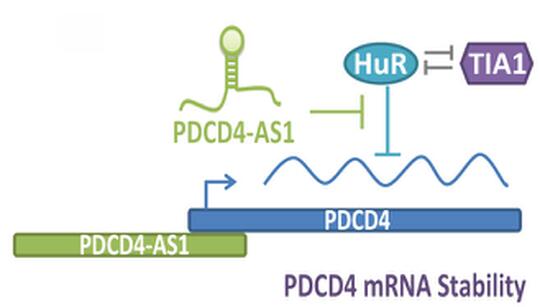
My laboratory is also keen to understand the molecular function of lncRNAs that are deregulated in BC, especially in triple-negative breast cancer (TNBC) or basal-like subtype, which is considered to be the most aggressive subtype among all of the BC sub-groups. We recently performed genome-wide transcriptome analyses in isogenic, TNBC/basal-like progression cell lines using a 3D cell culture model (Jadaliha et al., PloS Genetics 2018). By this, we identified aberrant expression of >1800 lncRNAs, through this transformation progression (Fig. 3). Functional characterization of one such lncRNA; PDCD4-AS1 revealed that it controls cancer progression by promoting the mRNA stability of its sense partner, the well-known tumor suppressor PDCD4. We are interested in, characterizing the biological function of several of the other lncRNAs that show deregulated expression in the isogenic BC model as well as in TNBC patients. High-throughput approaches such as CRISPRa- and CRISPRi-based genome editing will be employed for loss- or gain-of-function studies to address the role of candidate lncRNAs in controlling key cancer cell properties. Further, the roles of lncRNAs in activating or repressing key oncogenes or tumor suppressor genes will be determined. Our proposed studies will expand the knowledge on the role of lncRNAs in cancer cells, and detailed understanding of their role in cell cycle and cancer progression could potentially help to design strategies to control cancer proliferation, tumor progression and metastasis, opening an lncRNA-focused avenue for therapy.
The past decade has witnessed spectacular progress in the field of ncRNA biology. We have seen an explosion in the number of identified lncRNAs, yet we have only begun to appreciate the complexity of lncRNAs, and how cells utilize this class of ‘riboregulators’ for various aspects of gene expression. The most challenging and exciting areas of future research lies in the functional characterization of the roles played by several of these lncRNAs. In the next several years, we will work towards unraveling the role played by lncRNAs in the proper functioning of eukaryotic cells, their role in regulating vital gene regulatory pathways, and their involvement in cancer progression.
Education
M.Sc., Vector Control Research Center, Pondicherry, India
Ph.D., Cytogenetics Laboratory, Banaras Hindu University, Varanasi, India
Postdoctoral Fellow, Cold Spring Harbor Laboratory, New York, USA
Awards and Honors
EAGER NSF award
Research Scholar, American Cancer Society
Cancer Center at Illinois seed grants
Courses Taught
Additional Campus Affiliations
Professor, Cell and Developmental Biology
External Links
Highlighted Publications
Hao, Q., Liu, M., Daulatabad, S. V., Gaffari, S., Song, Y. J., Srivastava, R., Bhaskar, S., Moitra, A., Mangan, H., Tseng, E., Gilmore, R. B., Frier, S. M., Chen, X., Wang, C., Huang, S., Chamberlain, S., Jin, H., Korlach, J., McStay, B., ... Prasanth, K. V. (2024). Monoallelically expressed noncoding RNAs form nucleolar territories on NOR-containing chromosomes and regulate rRNA expression. eLife, 13, Article e80684. https://doi.org/10.7554/eLife.80684
Hao Q & Prasanth KV (2021) Regulatory roles of nucleolus organizer region-derived long noncoding RNAs. Mammalian Genome 2022 Jun;33(2):402-411. doi: 10.1007/s00335-021-09906-z..
Han et al., (2021) The BRCA1Pseudogene Negatively Regulates Anti-Tumor Responses through Inhibition of Innate Immune Defense Mechanisms. Cancer Research: 81(6):1540-1551. doi: 10.1158/0008-5472.CAN-20-1959.
Muys BR et al., (2021) The p53-induced RNA-binding protein ZMAT3 is a splicing regulator that inhibits the splicing of oncogenic CD44 variants in colorectal carcinoma. Genes and Dev. 35(1-2):102-116. doi: 10.1101/gad.342634.120.
Li et al., (2020) A small protein encoded by a putative lncRNA regulates apoptosis and tumorigenicity in human colorectal cancer cells. Elife. 2020 Oct 28;9:e53734. doi: 10.7554/eLife.53734.
Qinyu Hao*, Xinying Zong*, et al., (2020) The S phase-induced lncRNA SUNO1 promotes cell proliferation by controlling YAP1/Hippo signaling pathway. Elife. 2020 Oct 27;9:e55102. doi: 10.7554/eLife.55102.
Jadaliha M, et al., (2018) A Natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 14(11):e1007802. doi: 10.1371/journal.pgen.1007802.
Sun et al., (2018) MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs, Nucleic Acids Res. 2018 Aug 8. doi: 10.1093/nar/gky696. [Epub ahead of print].
Fei et al., (2017) Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super-resolution. J. Cell Science 2017 Dec 15; 130(24):4180-4192.
Li XL, et al., (2017) Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Reports 20(10): 2408-23.
Anantharaman A. et al., (2017) ADAR2 regulates RNA stability by modifying access of decay-promoting RNA-binding proteins. Nucleic Acids Res. Apr 20;45(7):4189-4201.
Malakar P et al., (2017) Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 up-regulation and mTOR activation. Cancer Res. Mar 1;77(5):1155-1167.
Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. (2013) Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. PLoS Genet. 9(3):e1003368.
Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., Blencowe B.J., Prasanth S.G. and Prasanth K.V. (2010) The Nuclear-retained Non-coding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Molecular Cell 39(6): 925-938.
Recent Publications
Hao, Q., Liu, M., Daulatabad, S. V., Gaffari, S., Song, Y. J., Srivastava, R., Bhaskar, S., Moitra, A., Mangan, H., Tseng, E., Gilmore, R. B., Frier, S. M., Chen, X., Wang, C., Huang, S., Chamberlain, S., Jin, H., Korlach, J., McStay, B., ... Prasanth, K. V. (2024). Monoallelically expressed noncoding RNAs form nucleolar territories on NOR-containing chromosomes and regulate rRNA expression. eLife, 13, Article e80684. https://doi.org/10.7554/eLife.80684
Arif, W., Mathur, B., Saikali, M. F., Chembazhi, U. V., Toohill, K., Song, Y. J., Hao, Q., Karimi, S., Blue, S. M., Yee, B. A., Van Nostrand, E. L., Bangru, S., Guzman, G., Yeo, G. W., Prasanth, K. V., Anakk, S., Cummins, C. L., & Kalsotra, A. (2023). Splicing factor SRSF1 deficiency in the liver triggers NASH-like pathology and cell death. Nature communications, 14(1), Article 551. https://doi.org/10.1038/s41467-023-35932-3
Emon, B., Song, Y. J., Joy, M. S. H., Kovour, M. V., Prasanth, K. V., & Saif, M. T. A. (2023). Mechanosensitive changes in the expression of genes in colorectal cancer-associated fibroblasts. Scientific Data, 10(1), Article 350. https://doi.org/10.1038/s41597-023-02233-9
Hsieh, P. H., Phal, Y., Prasanth, K. V., & Bhargava, R. (2023). Cell Phase Identification in a Three-Dimensional Engineered Tumor Model by Infrared Spectroscopic Imaging. Analytical Chemistry, 95(6), 3349-3357. https://doi.org/10.1021/acs.analchem.2c04554
Lin, Y. C., Liu, D., Chakraborty, A., Macias, V., Brister, E., Sonalkar, J., Shen, L., Mitra, J., Ha, T., Kajdacsy-Balla, A., Prasanth, K. V., & Prasanth, S. G. (2023). DNA Damage-Induced, S-Phase Specific Phosphorylation of Orc6 is Critical for the Maintenance of Genome Stability. Molecular and cellular biology, 43(4), 143-156. https://doi.org/10.1080/10985549.2023.2196204




