
Contact Information
MC-110
601 S. Goodwin Ave.
Urbana, IL 61801
Research Interests
Research Topics
Archaea, Bioinformatics, Genome Organization, Host-Pathogen Interactions, Microbial Ecology, Microbial Physiology, Molecular Evolution, Virology
Disease Research Interests
Infectious Diseases
Research Description
The Whitaker Lab studies the evolution of Archaea, Bacteria and Viruses in natural and clinical environments.
All who are paying attention now appreciate the complexity, connectivity, and interdependence of biological systems wherein a single 30kb piece of single-stranded RNA can have planetary impacts. It is now abundantly clear that all living organisms are connected to each other through hidden microbial networks, and that while these connections are highly localized, microbial meta-populations act globally. Solving the global challenges of infectious disease, antibiotic resistance, and stabilization of healthy microbial ecosystems depends upon understanding the evolutionary dynamics of microbes and co-evolution of microbes and their own infectious elements. The Whitaker Lab studies on the dynamics of microbes and their viruses using a combination of genomics, experimental evolution, modeling, and molecular biology in a diversity of systems to provide a comparative context for discovering foundational rules of infection genomics. We focus on understanding how the physiology and evolution of rapidly changing components of microbial genomes including CRISPR-Cas immunity and genomic islands change microbial interactions in their natural contexts.
Viral Symbiosis
Viruses are the next frontier of microbiology not only because they are often pathogens, but also because they are the infectious source of genome innovation. Our research explores how viruses that are integrated in their host’s genome confer new evolutionary traits to their hosts and change their eco-evolutionary interactions with other organisms. With funding from the Gordon and Betty Moore Foundation our lab studies how viral symbioses results in dynamics and eco-evolutionary feedbacks in highly structured, wild archaeal and viral populations from Yellowstone hot springs. We focus on terrestrial hot springs as an example of highly structured populations in which local adaptation and random processes will likely play an important role in shaping symbiotic interactions.
One Health
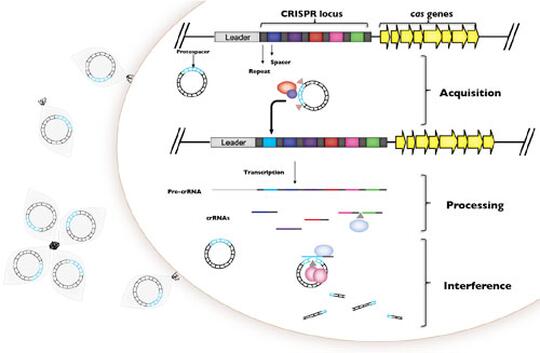
Viruses (phage) that infect bacterial pathogens such as P. aeruginosa are a key determinant of virulence, pathogenicity, epidemiology, adaptation, and evolution within and between the human host islands. We explore the transmission dynamics of P. aeruginosa phage within and between P. aeruginosa strains and how CRISPR-Cas immunity reflects the coevolutionary dynamics of P. aeruginosa and its viral populations. We extend this to other pathogen systems in which CRISPR-Cas systems are active to develop predictive models for the spread of virulence and antibiotic resistance in these systems. Through the Infection Genomics for One Health Theme at the Carl R. Woese Institute of Genomic Biology we extend the theoretical and molecular understanding of these systems further into a diversity of systems that connect observation to experiment across the continuum of symbiosis from antagonism to mutualism.
Archaeal Cell Biology
The movement of new genes and alleles into, and out of, microbial genomes defines rates of adaptation, population diversity, and genome architecture. Working in the only experimentally tractable crenarchaea (Sulfolobus) as our model system we are exploring the physiological and evolutionary basis of archaeal genome architecture including viruses, plasmids, and other mobile elements and their organization and integration into the multi-origin chromosome of the last genetically tractable common ancestor to eukaryotic cells.
Education
B.A, (Biology, SiSP), Wesleyan University, 1993
Ph.D., (Microbiology), University of California, Berkeley, 1998-2004
Postdoctoral Researcher, (Geomicrobiolgy), University of California, Berkeley, 2004-2006
Awards and Honors
Teachers Ranked as Excellent
Allen Distinguished Investigator Award (2017)
University Scholar Award (2020)
American Academy of Microbiology Fellow
Co-Director of the Microbial Diversity Course at the Marine Biological Lab in Woods Hole, MA
Additional Campus Affiliations
External Links
Co-editor of "Women in Microbiology"
Highlighted Publications
DeWerff, Samantha J., Maria A. Bautista, Matthew Pauly, Changyi Zhang, and Rachel J. Whitaker. “Killer Archaea: Virus-Mediated Antagonism to CRISPR-Immune Populations Results in Emergent Virus-Host Mutualism.” MBio 11, no. 2 (April 28, 2020). https://doi.org/10.1128/mBio.00404-20.
Rowland, Elizabeth F., Maria A. Bautista, Changyi Zhang, and Rachel J. Whitaker. “Surface Resistance to SSVs and SIRVs in Pilin Deletions of Sulfolobus Islandicus.” Molecular Microbiology 113, no. 4 (2020): 718–27. https://doi.org/10.1111/mmi.14435.
Pilosof, Shai, Sergio A. Alcala-Corona, Tong Wang, Ted Kim, Sergei Maslov, Rachel J. Whitaker, and Mercedes Pascual. “The Network Structure and Eco-Evolutionary Dynamics of CRISPR-Induced Immune Diversification.” BioRxiv, November 22, 2019, 850800. https://doi.org/10.1101/850800.
England, Whitney E., Ted Kim, and Rachel J. Whitaker. “Metapopulation Structure of CRISPR-Cas Immunity in Pseudomonas Aeruginosa and Its Viruses.” MSystems 3, no. 5 (October 30, 2018). https://doi.org/10.1128/mSystems.00075-18.
Westra, Edze R., Stineke van Houte, Sylvain Gandon, and Rachel Whitaker. “The Ecology and Evolution of Microbial CRISPR-Cas Adaptive Immune Systems.” Philosophical Transactions of the Royal Society B: Biological Sciences 374, no. 1772 (May 13, 2019): 20190101. https://doi.org/10.1098/rstb.2019.0101.
Zhang, Changyi, Alex P. R. Phillips, Rebecca L. Wipfler, Gary J. Olsen, and Rachel J. Whitaker. “The Essential Genome of the Crenarchaeal Model Sulfolobus Islandicus.” Nature Communications 9, no. 1 (December 2018): 4908. https://doi.org/10.1038/s41467-018-07379-4.
Zhang, Changyi, Rebecca L. Wipfler, Yuan Li, Zhiyu Wang, Emily N. Hallett, and Rachel J. Whitaker. “Cell Structure Changes in the Hyperthermophilic Crenarchaeon Sulfolobus Islandicus Lacking the S-Layer.” MBio 10, no. 4 (August 27, 2019): e01589-19. https://doi.org/10.1128/mBio.01589-19.
Recent Publications
Collins, A. J., & Whitaker, R. J. (2023). CRISPR Comparison Toolkit: Rapid Identification, Visualization, and Analysis of CRISPR Array Diversity. CRISPR Journal, 6(4), 386-400. https://doi.org/10.1089/crispr.2022.0080
Kim, H. W., Kim, N. K., Phillips, A. P. R., Parker, D. A., Liu, P., Whitaker, R. J., Rao, C. V., & Mackie, R. I. (2023). Genomic insight and physiological characterization of thermoacidophilic Alicyclobacillus isolated from Yellowstone National Park. Frontiers in Microbiology, 14, Article 1232587. https://doi.org/10.3389/fmicb.2023.1232587
Kosmopoulos, J. C., Campbell, D. E., Whitaker, R. J., & Wilbanks, E. G. (2023). Horizontal Gene Transfer and CRISPR Targeting Drive Phage- Bacterial Host Interactions and Coevolution in "Pink Berry" Marine Microbial Aggregates. Applied and environmental microbiology, 89(7). https://doi.org/10.1128/aem.00177-23
Mao, Y., Zeineldin, M., Usmani, M., Jutla, A., Shisler, J. L., Whitaker, R. J., & Nguyen, T. H. (2023). Local and Environmental Reservoirs of Salmonella enterica After Hurricane Florence Flooding. GeoHealth, 7(11), Article e2023GH000877. https://doi.org/10.1029/2023GH000877
Sanchez-Nieves, R. L., Zhang, C., & Whitaker, R. J. (2023). Integrated conjugative plasmid drives high frequency chromosomal gene transfer in Sulfolobus islandicus. Frontiers in Microbiology, 14, Article 1114574. https://doi.org/10.3389/fmicb.2023.1114574




