
Contact Information
Research Interests
Research Topics
Genetics, Genomics, Metabolic Regulation, Microbial Physiology, Regulation of Gene Expression, RNA Biology, Signal Transduction
Research Description
The Vanderpool lab investigates RNA-mediated mechanisms of genetic regulation in bacteria.
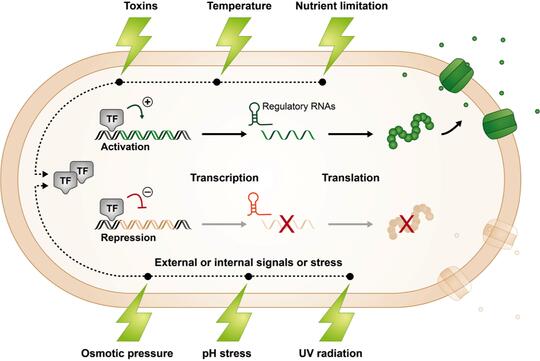
Bacteria living in natural environments must adjust their gene expression in response to changing, and often stressful, conditions so that they can continue to grow and compete with their neighbors for limited resources.
While bacteria employ different strategies to regulate gene expression, our lab is interested in a type of regulation involving small RNAs (sRNAs). sRNAs are typically non-coding RNA molecules that can be found in organisms belonging to all domains of life.
In bacteria, they are often involved in regulating stress responses using mechanisms that involve base pairing with a target mRNA and enhancing or repressing its stability or translation. We are interested in deciphering the overall impact of sRNAs on cell physiology, metabolism, and structure and function.
Global discovery and characterization of small RNA regulatory networks
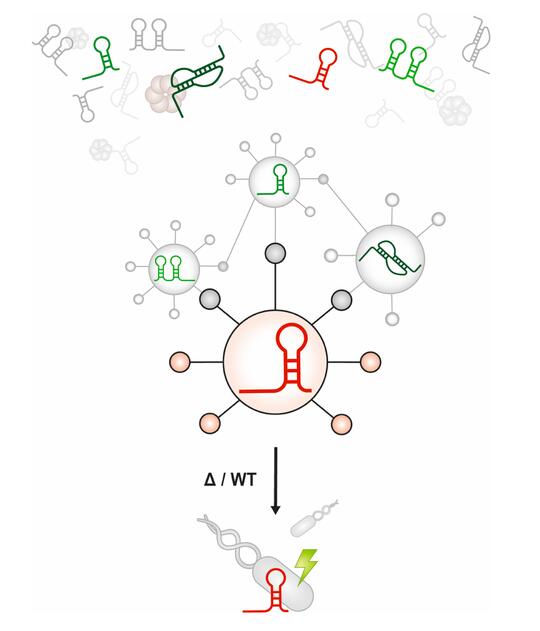
Bacterial genomes encode tens to hundreds of small RNAs (sRNAs), and we know little or nothing about their functions in most organisms.
Discovering the set of mRNA targets that a given sRNA regulates – the sRNA’s “regulon” – helps us begin to understand sRNA function. This is a difficult problem. Although a variety of computational and experimental methods exist for sRNA target identification, the process is labor-intensive and often yields a high false-positive rate.
We have attempted to improve the efficiency of sRNA target identification by developing and testing SPOT: sRNA Target Prediction Organizing Tool. This is a pipeline that can integrate computational predictions and experimental evidence to generate sRNA target regulon predictions. SPOT has so far helped us identify new target mRNAs for several sRNAs in E. coli.
We will use our results to further refine SPOT and define sRNA target regulons in other bacteria. Our ultimate goal is to have a comprehensive understanding of the roles of sRNAs in bacterial physiology and metabolism.
Post-transcriptional control of carbohydrate transport and metabolism
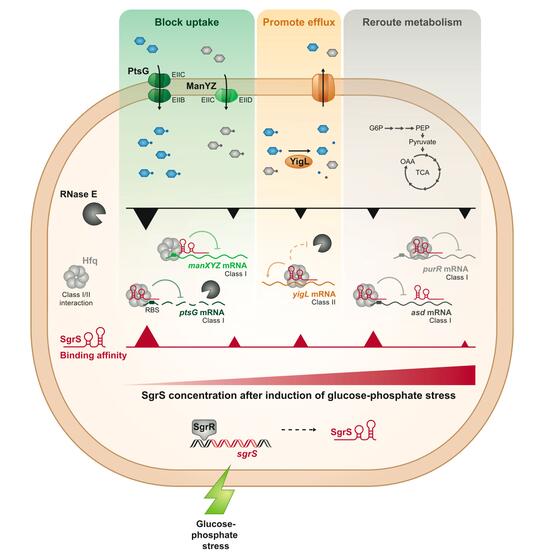
Our work on the sRNA SgrS has allowed us to develop one of the top existing models for discovery of new molecular mechanisms of regulation by bacterial sRNAs and sRNA-mediated control of bacterial metabolism and physiology.
SgrS (sugar stress sRNA) is produced by enteric bacteria like E. coli and Salmonella during metabolic stress conditions, when sugar metabolism can’t keep up with sugar transport. SgrS regulates nine genes to restore metabolic homeostasis and allow stressed cells to recover and continue growing. SgrS represses translation of ptsG and manXYZmRNAs, encoding sugar transporters, to temporarily block uptake of sugars. SgrS enhances translation of the yigL (sugar phosphatase) mRNA to promote dephosphorylation of sugars and allow them to be pumped out of the cell. SgrS regulates other targets to reroute metabolism around the metabolic block. SgrS prioritizes target regulation in a specific order so that cells rapidly and efficiently adapt to the stress and continue growing.
Small RNA-mediated control of membrane structure and function
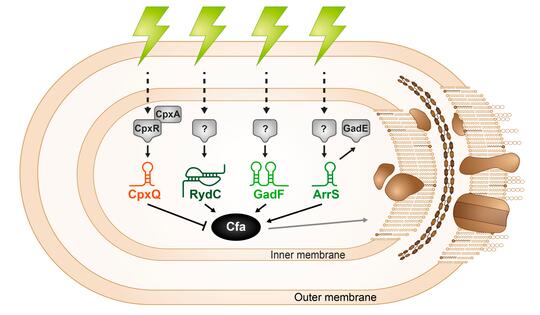
Microbial membranes are the first line of defense against environmental stress as well as the site of many metabolic processes, so membrane integrity is key to cell survival. Bacteria control the biophysical properties of membranes using lipid modifications like cyclopropanation. The cyclopropane fatty acid synthase (Cfa) enzyme catalyzes this lipid modification and is regulated in response to stress. The sRNA RydC was previously shown to stabilize cfa mRNA, resulting in higher levels of cyclopropane fatty acids in the cell membrane. We found additional activating (ArrS) and repressing (CpxQ) sRNAs that also directly regulate cfa post-transcriptionally. RydC and CpxQ exhibit pH-dependent regulation of cfa, and RydC-dependent cfa activation protects cells from acid shock. This work illustrates how mRNAs can serve as hubs for signal integration via multiple sRNAs responding to different environmental signals. Future work aims to understand the signals that induce production of each sRNA and more conditions where this sRNA-mediated regulation is beneficial for bacterial cells.
Prophage-encoded regulators control bacterial host metabolism and physiology
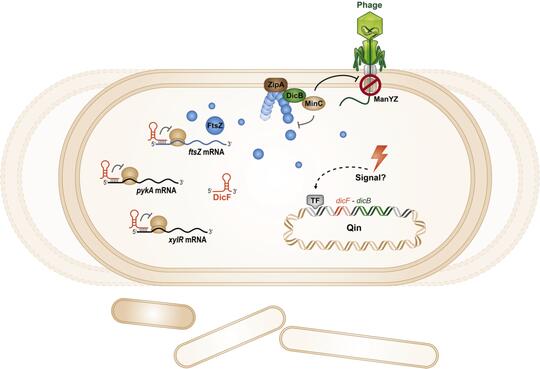
Cryptic prophages, which have lost the ability to produce phage progeny, have been discovered within bacterial genomes and harbor genes of unknown function which impact the host cell physiology. We have characterized the sRNA DicF and associated small protein DicB within the cryptic prophage Qin in E. coli K12. DicF and DicB were known to be inhibitors of cell division. We found that DicF sRNA regulates mRNAs encoding cell division and metabolic activities via direct sRNA-mRNA base pairing. We also discovered a new function for the small protein DicB. Transient production of DicB protected E. coli cells from infection by lambda phage. DicB specifically provided protection against phages that use a specific cytoplasmic membrane receptor – ManYZ – for injection of their DNA into the E. coli cytoplasm. Our results suggest that one small prophage-encoded protein can carry out multiple functions that affect bacterial cell division and physiology. Our future work will aim to uncover the molecular mechanisms by which DicB carries out its multiple activities.
Genetics and physiology of gut microbes
Complex communities of microbes in our gut have tremendous impacts on our health by contributing metabolic functions, protecting against disease-causing pathogens, educating the immune system, controlling obesity, and even affecting learning and memory. We are interested in understanding the regulatory mechanisms that govern physiology and metabolism of important members of the gut microbiota belonging to the phylum Bacteroidetes. In these organisms, we know very little about RNA-mediated mechanisms of regulation. Work in our laboratory is focused on identification and characterization of novel RNA binding proteins and sRNAs.
Education
B.S. Microbiology, Purdue University, 1998
Ph.D. Microbiology, Immunology and Cancer Biology, University of Minnesota, 1998-2003
Postdoctoral Fellow, National Cancer Institute, 2003-2006
Awards and Honors
2018 Charles G Miller Professorial Scholar
2013 Helen Corley Petit Scholar, College of Liberal Arts and Sciences
Additional Campus Affiliations
External Links
Highlighted Publications
Representative Publications
Bobrovskyy, M., Azam, M.S., Frandsen, J.K., Zhang, J., Poddar, A., Ma, X., Henkin, T.M., Ha, T., and Vanderpool, C.K. 2019. Determinants of target prioritization and regulatory hierarchy for the bacterial small RNA SgrS. Mol. Microbiol.
Bianco, C.M., Fröhlich, K.S., and Vanderpool, C.K. 2019. Bacterial Cyclopropane Fatty Acid Synthase mRNA is targeted by activating and repressing small RNAs. J. Bacteriol.
King, A.M., Vanderpool, C.K., and Degnan, P.H. 2019. sRNA Target Prediction Organizing Tool (SPOT) Integrates Computational and Experimental Data To Facilitate Functional Characterization of Bacterial Small RNAs. mSphere 4.
Kim, K., Palmer, A.D., Vanderpool, C.K., and Slauch, J.M. 2019. The sRNA PinT contributes to PhoP-mediated regulation of the SPI1 T3SS in Salmonella enterica serovar Typhimurium. J. Bacteriol.
Kim, K., Golubeva, Y.A., Vanderpool, C.K., and Slauch, J.M. 2019. Oxygen-dependent regulation of SPI1 type three secretion system by small RNAs in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 111:570–587.
Azam, M.S., and Vanderpool, C.K. 2018. Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res. 46:2585–2599.
Lloyd, C.R., Park, S., Fei, J., and Vanderpool, C.K. 2017. The Small Protein SgrT Controls Transport Activity of the Glucose-Specific Phosphotransferase System. J. Bacteriol. 199.
Balasubramanian, D., Ragunathan, P.T., Fei, J., and Vanderpool, C.K. 2016. A Prophage-Encoded Small RNA Controls Metabolism and Cell Division in Escherichia coli. mSystems 1.
Bobrovskyy, M., and Vanderpool, C.K. 2016. Diverse mechanisms of post-transcriptional repression by the small RNA regulator of glucose-phosphate stress. Mol. Microbiol. 99:254–273.
Fei, J., Singh, D., Zhang, Q., Park, S., Balasubramanian, D., Golding, I., Vanderpool, C.K., and Ha, T. 2015. RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science 347:1371–1374.
Richards, G.R., Patel, M.V., Lloyd, C.R., and Vanderpool, C.K. 2013. Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J. Bacteriol. 195:4816–4825.
Balasubramanian, D., and Vanderpool, C.K. 2013. Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J. Bacteriol. 195:4620–4630.
Papenfort, K., Sun, Y., Miyakoshi, M., Vanderpool, C.K. and J. Vogel. 2013. Regulation of glucose homeostasis by small RNA mediated suboperonic mRNA stabilization. Cell 153:426.
Recent Publications
Abdulla, S. Z., Kim, K., Azam, M. S., Golubeva, Y. A., Cakar, F., Slauch, J. M., & Vanderpool, C. K. (2023). Small RNAs Activate Salmonella Pathogenicity Island 1 by Modulating mRNA Stability through the hilD mRNA 39 Untranslated Region. Journal of bacteriology, 205(1). https://doi.org/10.1128/jb.00333-22
Ragunathan, P. T., Lim, E. N. K., Ma, X., Massé, E., & Vanderpool, C. K. (2023). Mechanisms of Regulation of Cryptic Prophage-Encoded Gene Products in Escherichia coli. Journal of bacteriology, 205(8). https://doi.org/10.1128/jb.00129-23
Cakar, F., Golubeva, Y. A., Vanderpool, C. K., & Slauch, J. M. (2022). The Small RNA MicC Downregulates hilD Translation to Control the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium. Journal of bacteriology, 204(1), Article e00378-21. https://doi.org/10.1128/JB.00378-21
Adams, A. N. D., Azam, M. S., Costliow, Z. A., Ma, X., Degnan, P. H., & Vanderpool, C. K. (2021). A novel family of RNA-binding proteins regulate polysaccharide metabolism in bacteroides thetaiotaomicron. Journal of bacteriology, 203(21), Article e00217-21. https://doi.org/10.1128/JB.00217-21
Lalaouna, D., Prévost, K., Park, S., Chénard, T., Bouchard, M.-P., Caron, M.-P., Vanderpool, C. K., Fei, J., & Massé, E. (2021). Binding of the RNA Chaperone Hfq on Target mRNAs Promotes the Small RNA RyhB-Induced Degradation in Escherichia coli. Non-coding RNA, 7(4), Article 64. https://doi.org/10.3390/ncrna7040064




