
Contact Information
B303 CLSL
601 S Goodwin Ave
Urbana, IL 61801
Research Interests
Research Topics
DNA Biology, Drug Discovery, Enzymology, Microbial Ecology, Microbial Physiology
Research Description
Molecular mechanisms of oxidative damage; cellular defenses against oxidants; obligate anaerobiosis
Oxygen is a reactive chemical, and its gradual accumulation in the environment must have been a problem for early life forms. For kinetic reasons much of its toxicity is mediated by its partially reduced forms, superoxide (O2-) and hydrogen peroxide (H2O2). Virtually all aerobic organisms contain enzymes devoted to eliminating these species from the cell: superoxide dismutases and reductases for O2-, and catalases and peroxidases for H2O2. This implies that O2- and H2O2 are formed somehow during aerobic metabolism and that, unless scavenged, they will harm the cell. Indeed, mutants that are unable to detoxify these oxygen species have difficulty growing in the presence of oxygen.
We investigate fundamental questions about O2- and H2O2 toxicity: How do these species arise in the cell? What damage can they do? What do cells do to avoid or repair that damage? And when in nature does oxidative damage determine the fate of the cell?
Our laboratory pursues these questions in a number of microbes but with a particular focus upon E. coli, a model organism whose metabolism is well-understood. However, the mechanisms by which oxidants damage E. coli, and the strategies by which E. colidefends itself, are widely shared by other bacteria—and, indeed, by higher organisms.
How are superoxide and hydrogen peroxide formed inside cells? Mutant strains of E. coli that lack superoxide dismutase or catalases/peroxidases are very sick. Yet E. coli has no enzymes that produce O2- or H2O2 as deliberate products. So what is the source of the O2- and H2O2 that disable these mutants? We have found that some flavoenzymes inadvertently transfer electrons to dissolved oxygen when it bounces into their reduced cofactors. O2- is formed if one electron is transferred, and H2O2 is formed if two electrons are transferred. Not surprisingly, this behavior is particularly marked if the flavin is solvent-exposed, has a high electron density, and has a low reduction potential. Several respiratory dehydrogenases have these traits, and they generate significant O2- and H2O2 during respiration. However, genetic analyses show that the primary source(s) of oxidants inside E. coli lie outside the respiratory chain. We have identified some of these sources; the identification of the others is an important goal of our lab.
What biomolecules does superoxide damage? Superoxide and hydrogen peroxide are used as weapons throughout the biological world: microbes and plants secrete antibiotics which, when ingested by competitors, redox-cycle to generate crippling doses of intracellular O2- and H2O2, and mammals blast invasive bacteria with O2- and H2O2 that is generated by specialized enzymes on the phagosomal membranes of neutrophils and macrophages. Yet, despite the fact that 100,000+ articles that address superoxide can be found in PubMed, we still are trying to identify the biological molecules that it damages. Superoxide does not react directly with polypeptides, sugars, or nucleic acids, and it is unlikely to trigger lipid peroxidation because bacterial membranes lack polyunsaturated fatty acids. Earlier studies in E. coli revealed that O2-disables the TCA cycle and branched-chain biosynthetic pathways by destroying the iron-sulfur clusters of critical enzymes. We have recently determined that O2- also inactivates certain enzymes that use a single ferrous iron atom to bind their substrates. Superoxide oxidizes this iron atom, the oxidized iron falls out of the enzyme, and other metals--that do not provide activity--bind in its place. This family of enzymes includes members that catalyze a wide range of reaction types, and their inactivation blocks a variety of cell functions. These must not be the only toxic actions of O2-, however, since SOD-deficient E. coli exhibit metabolic defects that these injuries cannot explain. We are analyzing these phenotypes in order to pinpoint additional biomolecules that O2-damages.
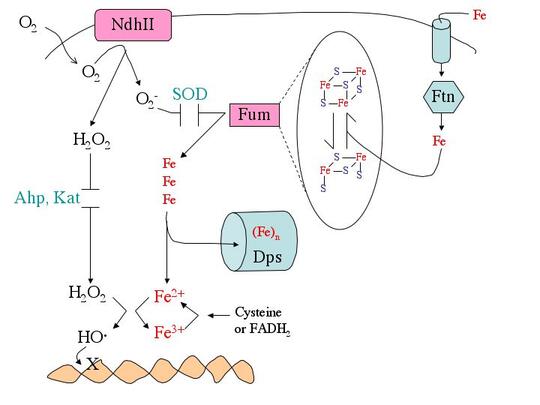
What biomolecules does hydrogen peroxide damage? H2O2 easily oxidizes reduced iron. This behavior has several consequences for cell health. First, like O2-, H2O2 directly damages the iron-sulfur clusters of key enzymes. Second, H2O2 also inactivates enzymes that use mononuclear Fe(II) as a catalytic cofactor. Third, H2O2reacts with the intracellular pool of unincorporated ferrous iron and thereby generates hydroxyl radicals. The latter species react avidly with virtually all biomolecules—including DNA. This injury causes mutations, a fact that has led many scientists to postulate that in mammals chronic oxidative DNA damage is a root cause of carcinogenesis and, perhaps, aging. Finally, as a consequence of its reactions with iron, H2O2 generally disrupts iron homeostasis and metabolism. This makes it difficult for cells to deliver iron into new metalloproteins. We are working to illuminate details of these damage processes (see figure).
How do microbes defend themselves against oxidants? While superoxide dismutase, catalases, and peroxidases scavenge O2- and H2O2, they can never diminish the intracellular levels to zero. Therefore many additional defensive measures are taken. E. coli repairs the iron-sulfur clusters of damaged enzymes by re-reducing their clusters and replacing their lost iron atoms. If the rate of enzyme damage exceeds the repair capacity, E. coli goes further, replacing some of the damaged iron-sulfur enzymes--e.g., fumarase—with isozymes that do not use iron and that are therefore resistant to oxidation. During periods of oxidative stress E. coli also synthesizes Dps, an iron-sequestering mini-ferritin that minimizes the amount of unincorporated iron that is available to react with H2O2. This action substantially suppresses DNA damage. Since this action would leave iron-dependent enzymes without a cofactor metal, the cell simultaneously activates the import of manganese as a replacement metal. Each of these adjustments is necessary—if any one of them is blocked, H2O2-stressed cells do not survive. Genetic analyses have identified many other genes that are deliberately induced in response to O2- or H2O2. Intriguingly, one of these enzymes is a respiration-coupled peroxidase that even allows microbes to exploit environmental H2O2 as a respiratory acceptor.
Why can't anaerobes grow aerobically? How do phagocytes kill bacteria? And other questions. . . . Oxidative stress determines the fate of bacteria in several situations. Obligate anaerobes cannot grow in the presence of oxygen, a feature that profoundly constrains their lifestyle and shapes microbial communities but is not well understood. Contrary to early models, anaerobes possess potent scavenging systems and inducible defenses. Using Bacteroides thetaiotaomicron as a model anaerobe, we determined that molecular oxygen itself impairs metabolism in several ways: by directly quenching radical-based enzymes, by oxidizing low-potential enzymic metal centers, and by triggering such rapid O2- formation that the superoxide dismutase is inadequate. This O2- inactivates the same classes of iron-sulfur and Fe(II) enzymes that it targets in E. coli. Yet we also found that metabolism soon resumes after anoxia is restored--indicating that this anaerobe has evolved effective ways of recovering from transient oxygen exposure. Incidental aeration must be a common event in many habitats, so we wish to dissect the recovery process.
Most bacteria are overpowered by the oxidative burst of phagocytes—but a select few pathogens somehow tolerate it. We know that some of the oxidative defenses that were identified in E. coli are essential to the success of invading pathogens. Thus the lessons from E. coli provide direction for investigations of oxidative stress in these other organisms as well.
Education
B.S. (Chemistry, English), Duke University, 1981
Ph.D. (Biochemistry), University of California, Berkeley, 1987
Postdoctoral (Biochemistry), Duke University, 1987-1992
Additional Campus Affiliations
Associate Head, Microbiology
Professor, Microbiology
External Links
Highlighted Publications
Representative Publications
Lu, Zheng, and James A. Imlay. 2021. When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance, and defence. Nat. Rev. Microbiology. 19: 774-785.
Gupta, Anshika, and James A. Imlay. 2022. Escherichia coli induces DNA repair enzymes to protect itself from low-grade hydrogen peroxide stress. Mol. Microbiol. 117: 754-769.
Zhou, Yidan, and James A. Imlay. 2022. Escherichia coli uses a dedicated importer and desulfidase to ferment cysteine. mBio 13: e0296521.
Giannakis, Stefanos, Anshika Gupta, Cesar Pulgarin, and James A. Imlay. 2022. Identifying the mediators of intracellular E. coli inactivation under UVA light: the (photo)Fenton process and singlet oxygen. Water Research 221: 118740.
Rohaun, Sanjay K., and James A. Imlay. 2022. The vulnerability of radical SAM enzymes to oxidants and soft metals. Redox Biology 57: 102495.
Eben, Stefanie S., and James A. Imlay. 2023. Excess copper catalyzes protein disulfide bond formation in the bacterial periplasm but not in the cytoplasm. Mol. Microbiol. 119: 423-438.
Eben, Stephanie S., and James A. Imlay. 2023. Evidence that protein thiols are not primary targets of intracellular reactive oxygen species in growing Escherichia coli. Front. Microbiol., in press.
Gupta, Anshika, and James A. Imlay. 2023. How a natural antibiotic uses oxidative stress to kill oxidant-resistant bacteria. Proc. Natl. Acad. Sci. USA, in press.
Recent Publications
Eben, S. S., & Imlay, J. A. (2023). Evidence that protein thiols are not primary targets of intracellular reactive oxygen species in growing Escherichia coli. Frontiers in Microbiology, 14, Article 1305973. https://doi.org/10.3389/fmicb.2023.1305973
Eben, S. S., & Imlay, J. A. (2023). Excess copper catalyzes protein disulfide bond formation in the bacterial periplasm but not in the cytoplasm. Molecular Microbiology, 119(4), 423-438. https://doi.org/10.1111/mmi.15032
Gupta, A., & Imlay, J. A. (2023). How a natural antibiotic uses oxidative stress to kill oxidant-resistant bacteria. Proceedings of the National Academy of Sciences of the United States of America, 120(52), Article e2312110120. https://doi.org/10.1073/pnas.2312110120
Padron, G. C., Shuppara, A. M., Sharma, A., Koch, M. D., Palalay, J. J. S., Radin, J. N., Kehl-Fie, T. E., Imlay, J. A., & Sanfilippo, J. E. (2023). Shear rate sensitizes bacterial pathogens to H2O2 stress. Proceedings of the National Academy of Sciences of the United States of America, 120(11), Article e2216774120. https://doi.org/10.1073/pnas.2216774120
Giannakis, S., Gupta, A., Pulgarin, C., & Imlay, J. (2022). Identifying the mediators of intracellular E. coli inactivation under UVA light: The (photo) Fenton process and singlet oxygen. Water Research, 221, Article 118740. https://doi.org/10.1016/j.watres.2022.118740




