The image, a rendering of the primary visual cortex in a mouse, was awarded second place in the NIH’s “Show Us Your Brains” photo and video contest. The contest is sponsored by the NIH BRAIN Initiative (Brain Research Through Advancing Innovative Neurotechnologies), which launched in 2013 to develop and apply tools that precisely map and observe brain circuits. The annual contest celebrates the many research endeavors taking place across the country.
“Seeing is believing! Visualizing scientific data has been probably the most effective way of communicating information among scientists,” said Emad Tajkhorshid, the J. Woodland Hastings Endowed Chair of Biochemistry in the University of Illinois School of Molecular & Cellular Biology and the director of the NIH Center for Macromolecular Modeling and Bioinformatics.
“At Illinois, for three decades and with the support of NIH (P41-104601), a team of biophysicists and software developers has worked on making accurate, efficient, and easy-to-use tools for molecular scientists, now with hundreds of thousands of users around the world. The team is extending the scope of their powerful visualization capabilities into the realm of neuroscience, making it possible to look at and readily interact with the complex data sets produced by scientists investigating the brain,” Tajkhorshid said.
Modern neuroscience is undergoing a profound evolution due to systematic efforts that provide large, comprehensive datasets on properties of brain cells, their connectivity, and activity. Converting these widening streams of complex data into new knowledge via analysis and modeling is a major challenge for the field, he said.
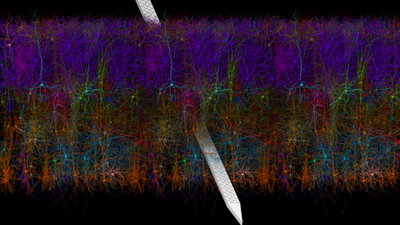
The image was created with new software developed at the NIH center. The group included software developers Barry Isralewitz, Mariano Spivak, and John Stone at the Beckman Institute for Advanced Science and Technology at the University of Illinois, in close collaboration with Anton Arkhipov and his team at the Allen Institute.
The software, called Visual Neuronal Dynamics, or VND, performs visualization of bio-realistic, 3D neuronal network models and aims to provide efficient workflows for large and complex systems of thousands of neurons and beyond. It is based on software previously developed by the group called Visual Molecular Dynamics, or VMD, a molecular visualization program for displaying, animating, and analyzing large biomolecular systems using 3D graphics and built-in scripting. The tool aims to make realistic models and simulation results of complex neuronal systems such as the brain accessible to all scientists through a few clicks. It not only provides an excellent research environment for neuroscientists, it also an excellent educational tool to communicate complex data to students and researchers from other disciplines.
NIH Director Francis Collins recently wrote about this powerful software in his blog.
