Congratulations to the University of Illinois scientists who were part of two teams selected as finalists for a prestigious award that celebrates outstanding achievement in high-performance computing.
The Association of Computing Machinery (ACM) Gordon Bell Special Prize for High Performance Computing–Based COVID-19 Research honors research achievements that advance the understanding of the COVID-19 pandemic using high-performance computing. Nominations were selected based on performance and innovation in their computational methods in addition to their contributions towards understanding the nature, spread and/or treatment of the disease.
Two simulation projects that included University Illinois researchers were selected as finalists for the prize.
“The Gordon Bell Prize, which is equated by some to the Nobel Prize in computing, recognizes achievements in applying our ever-increasing computational power to groundbreaking scientific or industrial problems. The U of I’s software programs, NAMD and VMD, which were specifically tuned in major ways to enable two of the finalists this year (and a winner last year!), were crucial to this success. We were thrilled that these two projects using NAMD and VMD were nominated for this award,” said Emad Tajkhorshid, J. W. Hastings Endowed Chair of Biochemistry, and director of the NIH Center for Macromolecular Modeling and Bioinformatics.
Team members from the University of Illinois provided performance and scalability advances in NAMD, the molecular dynamics simulation software developed by the Theoretical and Computational Biophysics Group (TCBG). NAMD already won a Gordon Bell Prize 20 years ago for its unparalleled achievement in parallel computing in biomolecular systems. They also provided technological improvements to VMD, a key molecular modeling tool used to prepare, visualize, and analyze molecular dynamics simulations of the aerosolized virus and spike protein. NAMD and VMD empowered scientists to more efficiently utilize state-of-the-art supercomputers, including Summit, the most powerful supercomputer in the United States, operated by Oak Ridge National Laboratory.
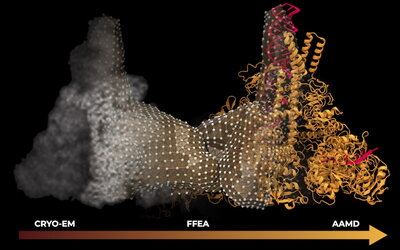
The first project nominated for the prize involved an unprecedented simulation of RNA unwinding in the SARS-CoV-2 replication transcription complex, which is the molecular "photocopier" responsible for viral replication inside human cells. Led by biophysics PhD students Anda Trifan and Defne Gorgun from Tajkhorshid's lab, in collaboration with Arvind Ramanathan's lab at Argonne National Laboratory, the researchers illuminated this crucial stage of infection by applying advanced features of the latest GPU-accelerated NAMD program. NAMD was developed by the NIH Center for Macromolecular Modeling and Bioinformatics, with John Stone, David Hardy, and James Philips as co-authors on the paper.
“Their involvement was super critical — both Anda and Defne exhibited a sense of perseverance and deep knowledge in using their skills to help us model the viral RNA moving through the ‘photocopier,’” Ramanathan said. “They brought together years of experience at UIUC and provided the critical momentum necessary for our project. Despite several personal challenges, both Anda and Defne dedicated their summer months to this project, which helped us move this project much farther along than where it was,” he said.
The team went beyond conventional simulation methodologies by coupling atomistic molecular dynamics simulations with other advanced mathematical models, namely, Fluctuating Finite Element Analysis (FFEA) and a hierarchy of deep learning methods that made sense of the intricate fluctuations in this molecular machine, according to Tajkhorshid.
The VMD software was extended to visualize for the first time FFEA simulations and to provide the team with unified views of all of the modalities of structure information and simulation trajectories used in the project. Together, Trifan and colleagues were able to provide invaluable insights into the function of the genetic photocopier while setting several high watermarks. The workflow coupled resources at Department of Energy facilities, with a novel wide-area data pipeline feeding intelligent agents on ThetaGPU with real-time simulations data from the supercomputer Perlmutter located across the country.
The other project, involving Anda Trifan, David Hardy, James Philips, and John Stone from the University of Illinois, in collaboration with multiple other institutions and led by Rommie Amaro’s lab at San Diego University, provided insights into the SARS-CoV-2 virus in an aerosol particle. Again, using NAMD, this is the first simulation of its kind breaking the billion atom mark.
Simulation of the SARS-CoV-2 virus, especially in its aerosolized form, has been an impossible task, but only until now. All-atom molecular dynamics simulation of supramolecular systems such as virtual structures have always presented tremendous challenges for researchers both in terms of their computation and their visualization. The massive size (one billion atoms) of the aerosolized virus model that was simulated with NAMD for the first time ever, and its complexity requiring innovative methods to construct a model for the simulation represent a prime example of what we can achieve these days towards understanding the molecular biology of the virion and its variants, Tajkhorshid said.
“The work of the NAMD/VMD teams was absolutely essential to the project as it’s the only software capable to construct, simulate, visualize and analyze the billion-atom aerosol simulations,” Professor Amaro said.
Their simulation work and the software that has powered the simulations have been highlighted in several media publications, including most recently, The New York Times.
More information on the projects can be found on the group's website and via the following videos:
