
As pathogenic bacteria play a game of cat and mouse with the human body’s immune system, the pathogens must adapt to changes in the environment to stay ahead of the immune system’s defenses. New findings by Illinois researchers challenge longstanding assumptions of how a particular family of enzymes that protect invaders and host cells alike from the universal threat of reactive oxygen species function and evolved. These enzymes, known as iron manganese superoxide dismutases, require either iron or manganse for function. In the context of infection, these enzymes play a critical role in protecting pathogens from the immune system. Their findings could help scientists learn to predict how pathogens adapt to our defenses, and in turn, aid in the development of counter-measures.
Thomas Kehl-Fie, a professor of microbiology in the School of Molecular & Cellular Biology, and collaborator Kevin Waldron, a professor at the Institute of Biochemistry & Biophysics in Warsaw, Poland, have found that have found that in response to the immune system restricting metal availability, the enzymes used by pathogens to survive have changed the metals that they use. The researchers published their findings in Nature Ecology & Evolution.
Through a combination of bioinformatics, microbiology, and structural biochemistry, they explored metal utilization of the iron manganese superoxide dismutases (SodFM) from across the tree of life. This analysis revealed changes in the use of iron and manganese for catalysis throughout this family’s evolution. Their findings also showed the SodFM family’s preferences shouldn’t be thought of as strict absolutes, but rather a spectrum of preferences along which enzymes can slide back and forth, Kehl-Fie said.
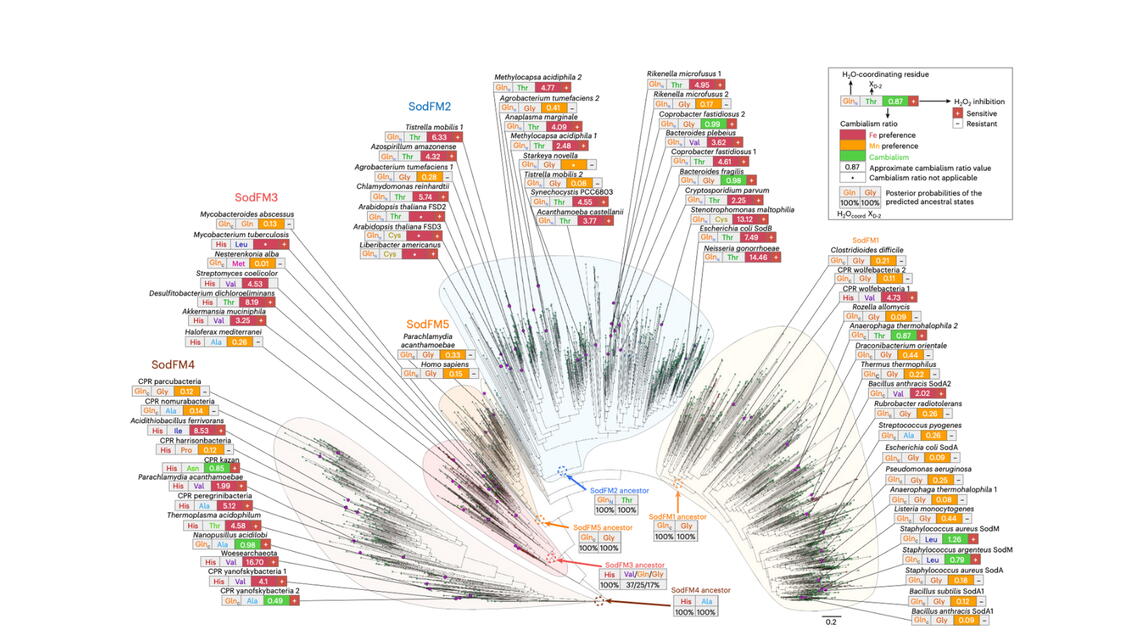
By employing bioinformatic analysis, Kehl-Fie and Waldron’s labs identified cases where the metal-preference of a SodFM enzyme changed during the evolution of a specific biological lineage. Although the two labs had previously detected a change in Staphylococcus aureus in 2020, the latest line of research confirmed that this was not an isolated evolutionary event.
“We tend to think of enzymes in their present state: ‘here they are and they exist in this way,’” Kehl-Fie explained. “But we saw that in modern-day organisms, there are frequent changes in metal utilization. This tells us that these enzymes are continually changing, enabling the microbes that produce them to adapt to new environments.”
A prime example of microbes shifting preferences is Staphylococcus aureus, a human bacterial pathogen that can cause serious infections or diseases, including endocarditis, meningitis and toxic shock syndrome. Mycobacterial pathogens, which can cause life-threatening illnesses such as tuberculosis, and major components of the microbiota also deploy such a change.
Kehl-Fie and Waldron’s lab observed SodFM’s metal preference switched multiple independent times throughout their research. Waldron voiced surprise at how flexible metalloenzymes were in their metal-preference.
“Whereas they were previously thought to be mostly of fixed specificity, having very strong preference for either manganese or iron, and with the cambialistic enzymes [can function equally well with either a manganese or iron cofactor] being rather rare quirks of chemistry, our data shows that this is actually not the case,” Waldron said. “Sampling a diverse range of enzymes from the SodFM family has demonstrated that their metal-preferences actually lie all along a sliding scale between the rare extremes of strong iron-preference and strong manganese-preference.”
Kehl-Fie and Waldron have spent the past decade collaborating in a study of how bacterial pathogens adapt. They’ve particularly focused on SodFM because of its presence in roughly 75% of all life on the planet. Through their latest research, they identified a new group of SodFMs, including several novel cambialistic enzymes that they plan to study in the future.
“By continuing to understand how metalloenzymes function and how metalloenzyme evolution happens in response to our immune system we will develop a greater understanding of how microbes transition from benign commensals to successful pathogens that threaten our health,” Kehl-Fie concluded. “Beyond this, metalloenzymes play critical roles in nearly all biological process and thus understanding how they function and can be adapted to new environments is critical for engineering organisms to benefit humanity.”