Sodium ions (Na+) play diverse and important roles in biological processes, and yet few sensors with high sensitivity and selectivity for Na+ over other competing metal ions have been reported. In this study, the authors reported the first highly selective, sensitive, and efficient Na+-specific catalytic DNA and its conversion into a sensor for imaging Na+ in living cells. Their findings have recently been published in PNAS.
During the past two decades, enormous progress has been made in designing fluorescent sensors for divalent metal ions such as Ca2+ and Zn2+. In contrast, the design of highly sensitive and selective fluorescent sensors for monovalent metal ions such as Na+ has been a major challenge for a long time, even though Na+ is the most abundant metal ion in biological systems, playing a critical role in diverse biological processes by triggering activation of signal transduction pathways, as well as influencing the actions of hormones. Among the limited number of reported Na+ sensors, most of them are not selective against K+ or have low binding affinity for Na+. Furthermore, the presence of organic solvents is required to achieve desired sensitivity and selectivity for many of the Na+ sensors, making it difficult to study Na+ under physiological conditions. We have advanced this field of monovalent ion detection by in vitro selecting a Na+-specific DNAzyme, called NaA43 DNAzyme, with high affinity and under physiological condition.
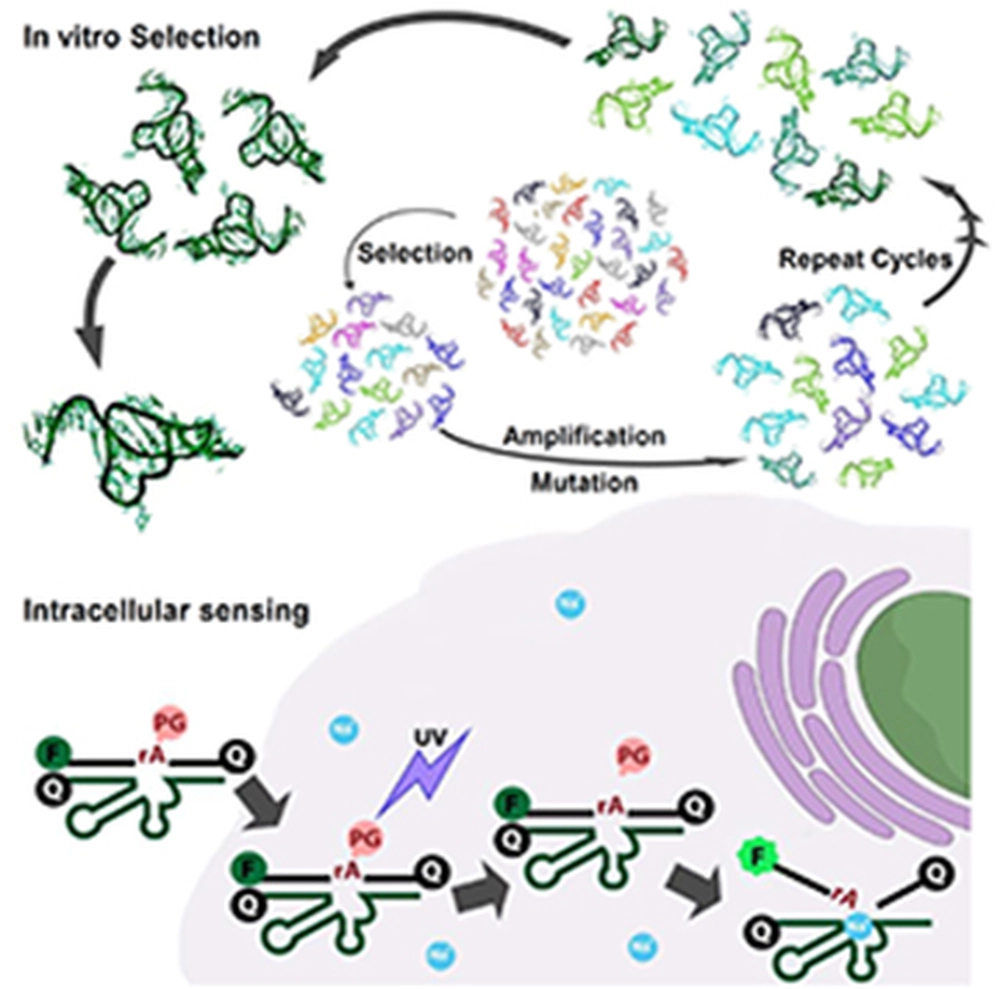
DNA which is known as a passive carrier of genetic information has been shown to have catalytic activity mostly in the presence of metal ions. Catalytic DNA (deoxyribozymes or DNAzymes) have been demonstrated to be a promising approach for detecting metal ions. However, most reported DNAzymes require multivalent metal ions for catalytic activity, and those DNAzymes that are active with monovalent ions typically have very poor selectivity for Na+ (~1.3 fold), require very high concentrations (molar range) for function and display low catalytic rate (10-3 min-1). We used a high throughput combinatorial approach called in vitro selection to isolate the first highly selective (>10,000 fold), sensitive (0.135 mM) and quickly-reacting (kobs ~ 0.1 min-1) RNA-cleaving Na+-specific DNAzyme (NaA43). The DNAzyme is then converted into a catalytic beacon fluorescent sensor for Na+. This Na+-specific DNAzyme was transformed into a DNAzyme-based fluorescent sensor and used in imaging intracellular Na+ in living cells, by employing an efficient DNAzyme delivery method using a cationic polypeptide, together with a photo-caging strategy to allow controllable activation of the Na+-specific DNAzyme-based probe inside cells with high spatial and temporal resolution.
The present study demonstrates for the first time a Na+-specific DNAzyme that can be used in living cells to monitor changes in the concentration of Na+. These findings open-up the possibility of addressing highly challenging questions related to the intracellular processes that are regulated by Na+. In addition, the results indicate that the NaA43 DNAzyme has a remarkable selectivity against K+, and other monovalent, divalent, and trivalent metal ions. In the protein world, although K+ channels are quite selective for K+ over other metal ions, it is known that the Na+ channels are much less selective. Therefore, obtaining such a highly Na+-selective DNAzyme not only provides an efficient sensor for Na+, it will give us the opportunity to investigate the origin of this unprecedented selectivity. We have performed further biochemical and biophysical studies to elucidate mechanism of Na+-selectivity in the NaA43 DNAzyme and will report the results in a future publication.
The National Institutes of Health supported this research.
Dr. Torabi is currently a postdoctoral fellow at Yale School of Medicine in Dr. Joan Steitz's group where he is conducting research on RNA motifs that stabilize non-coding RNA in cells.