A recent paper in PLOS ONE from the Kuzminov lab illustrated a novel phenomenon, RiCF, aka “RNase-induced chromosomal fragmentation.”
They report that degradation of RNA during lysis of Eschericha coli cells unexpectedly breaks the bacterial chromosome.
The research group has been studying DNA damage resulting from erroneous DNA metabolism and consequent chromosomal breakage. This chromosomal fragmentation is of particular interest, because genomic instability resulting from chromosomal breakage and repair has been implicated in various forms of cancer and neurodegenerative disease.
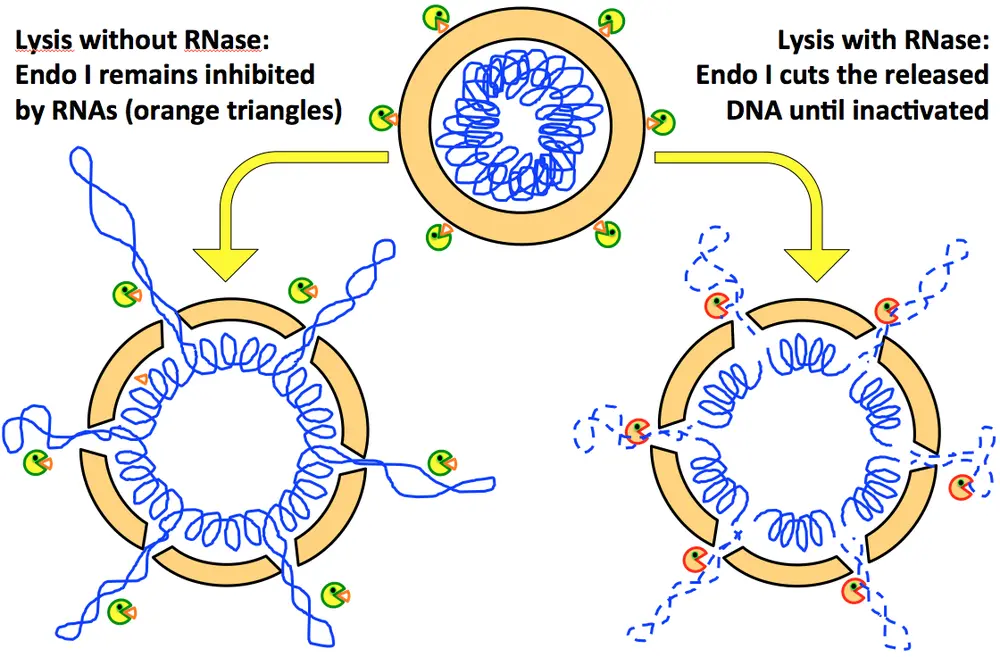
E. coli chromosome (nucleoid) comprises DNA, nucleoid-associated proteins (NAPs) and RNA with unknown function. In order to study double-strand breaks (DSBs) in chromosomal DNA, the researchers used genetically-tractable E. coli cells. To quantify DSBs, the investigators broke open (lysed) radiolabeled E. coli cells in agarose plugs and separated circular chromosomes from linear subchromosomal fragments using pulsed field gel electrophoresis (PFGE). This technique, in conjunction with restriction digestion, is typically used to obtain a DNA fingerprint from bacterial genome. However, when detecting chromosomal fragmentation, the isolated DNA in plugs is not digested, and if otherwise intact, stays in the wells. Conversely, broken DNA enters the gel to form a characteristic smear. While standardizing cell lysis and PFGE conditions, the researchers discovered massive chromosomal fragmentation if RNA degrading enzymes (RNases) were included in plugs during lysis. RNase-mediated changes in the released chromosomal content were expected considering the assumed role of RNA in the nucleoid structure and stability, and the massive RNase-triggered chromosomal fragmentation in lysing cells seemed to confirm the existence of a structural RNA organizing the nucleoid.
However, measuring RiCF in various mutants in the nucleoid-associated proteins (with the hope to find the one that interacts with the "nucleoid-organizing RNA") turned empty-handed, with a possible exception of H-NS. The investigators then looked for any potentially-DNA-cutting enzyme that was known to be inhibited by RNA. Eventually, such an enzyme was identified, but, surprisingly, it was not a part of the nucleioid!
The investigators showed that RiCF is completely dependent on an obscure enzyme endonuclease I, residing on the surface of bacterial cells and in the periplasm. The next interesting questions to explore are why would an extracytoplasmic nuclease always be inhibited with cellular RNA and what is its importance for bacterial cells.
