Update: The article has been featured by an invited commentary in PNAS.
From the commentary by Jana Shen:
“In PNAS, Vermaas et al. report a computational study that combines a battery of state-of the-art modeling and simulation tools to shed new light on the perplexing drug–proton antiport mechanism of EmrE at the atomic level.”
"The work of Vermaas et al. demonstrates the power of state-of-the-art modeling and molecular simulations in advancing our understanding of the structure–function relationships of membrane proteins that remain highly challenging to delineate with wet-laboratory experiments.”
Each year, at least 23,000 people die from infections caused by antibiotic resistant bacteria in the U.S., according to the Centers for Disease Control and Prevention. Using computer modeling, researchers from the University of Illinois at Urbana-Champaign and Sandia National Laboratories are helping to develop the means to prevent some of those deaths.
One way bacteria develop resistance to many different antibiotics is by producing pumps that spit out unfamiliar small molecules, such as antibiotics, before they can do the desired damage to bacteria. The researchers teased out the details of how one antibiotic pump works. This research was recently published in the Proceedings of the National Academy of Sciences.
The eventual goal is to develop new drugs to plug the pump so it cannot spit out antibiotics, perhaps restoring their effectiveness.
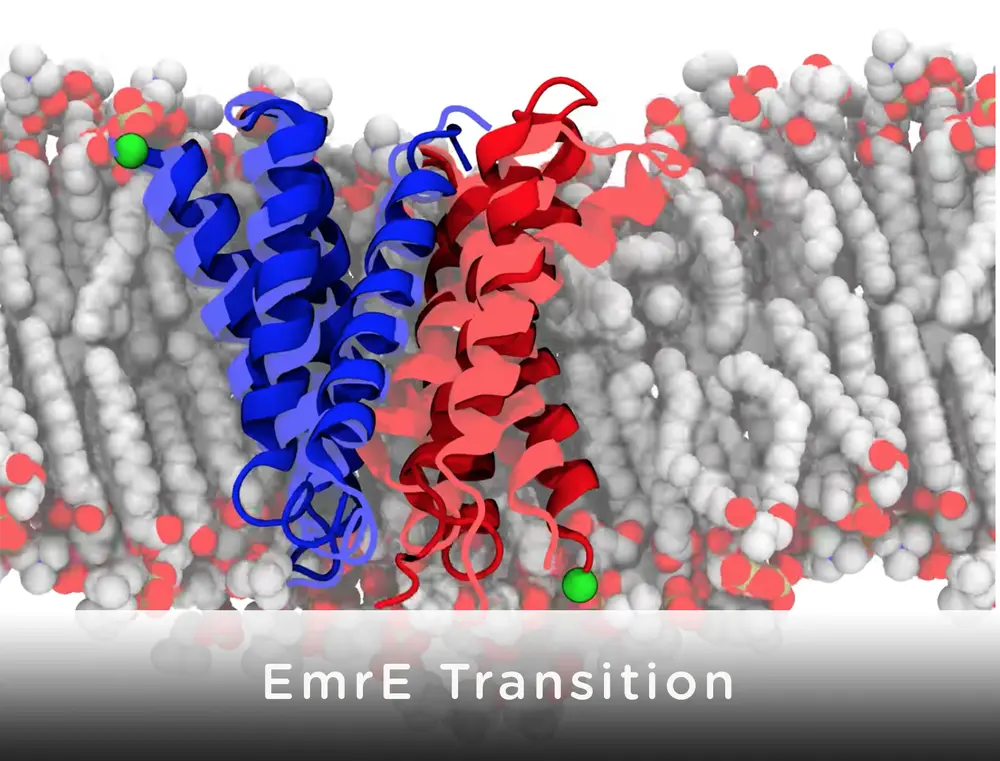
The specific pump researchers studied, called EmrE, comes from E. coli, common laboratory bacteria that can occasionally cause food poisoning. The pump recognizes and removes moderately oily, positively charged small molecules, said Josh Vermaas, a former biophysics graduate student at Illinois whose work under supervision of Emad Tajkhorshid (Biochemistry, Illinois) and Susan Rempe (Sandia National Labs) was supported through Sandia’s Campus Executive Program. Many common antibiotics including streptomycin, doxycycline and chloramphenicol are oily and positively charged and thus are expelled from the cell by this bacterial pump.
“Key to successful rational design of novel and effective inhibitors for EmrE, or any other transporter for that matter, is to describe the transport cycle of the transporter at an atomic resolution, that is, to understand how structural changes of these molecular machines mediate the movement of the drug molecule from the inside of the cell to the outside,” said Tajkhorshid, Endowed Professor of Biochemistry and Biophysics at Illinois. Their first step toward this goal was to determine a detailed structure of the pump. The existing, experimentally-determined structure of the pump was very rough, missing much of the essential chemical details, and misshapen, Vermaas said. It can be particularly challenging to get good structural data of drug transporters like EmrE because they are flexible.
Imagine having to take a picture of a wriggling toddler with a sluggish camera, the resulting photo is more of a blur than an exact likeness. They combined experimental data from a variety of common biophysical methods such as X-ray crystallography, cryo-electron microscopy (cryo-EM) and electron paramagnetic resonance (EPR) spectroscopy as well as decades of knowledge of the most likely internal arrangements of amino acids, the building blocks of proteins, to produce a high-resolution structure of the pump.
Once they had the detailed structure of the pump, the real work began.
First, the team added a lipid membrane to model the real-world environment of the pump. Then, they ran computer simulations to see what the pump looks like with zero, one or two protons. Letting in two protons is the battery that powers this pump. They ran simulations to see the transition from the protein facing inside the bacterium to outside in order to find the “easiest” path and thus see how the pump works. Modeling this “flip” took over 80,000 hours of computer processing.
They also ran simulations to see what the pump looks like with an example drug in the drug-binding pocket. Rempe said they found lots of flexibility in the pocket where antibiotics would bind, which makes sense given the pump can recognize a wide variety of drugs. They also identified a few critical amino acids that serve as a lock to make sure that the pump doesn’t let go of the protons willy-nilly.
“Antibiotic resistance is an important problem. The ‘lock’ on the pump is what makes this transporter tick. With this knowledge, in the future we can develop new antibiotics that aren’t pumped out or otherwise break the lock in EmrE,” said Vermaas. “If we figure out how to break the pump so it’s unregulated and leaks out protons, that would be a new way to kill bacteria.”
This project made extensive use of Sandia’s high-performance computing resources and the simulations were conducted at the Center for Integrated Nanotechnologies, a U.S. Department of Energy Office of Science user facility jointly operated by Sandia and Los Alamos National Laboratory. Sandia’s Laboratory Directed Research and Development office through the Campus Executive Program and the National Institutes of Health provided funding for the research. The Defense Threat Reduction Agency’s Joint Science and Technology Office provided additional funding.
Adapted from Sandia National Laboratories news release by Mollie Rappe.
