In biology, it is generally believed that a protein’s sequence determines its structure, which in turn determines its function. However, in the case of membrane proteins, the reality is more complicated than this simple statement. “In recent years, the importance of a protein’s local environment has been increasingly recognized, but the molecular-level details have not been quite clear”, says Dr. Yuhang (“Steven”) Wang, co-author of a recent paper from the Grosman and Tajkhorshid labs published earlier this month in PNAS.
In this work, the authors investigated the role of membrane lipids on a bacterial ion-channel membrane protein called ELIC, which shares the same protein architecture as the nicotinic acetylcholine receptors, GABA receptors, glycine receptors, and serotonin receptors found in the central and peripheral nervous systems of animals. The natural membrane environment of ELIC includes a variety of lipid species. When replacing this native membrane with one consisting of a single lipid species called POPC, the team found that the channel’s ion-conducting function is markedly reduced as indicated by electrophysiology experiments.
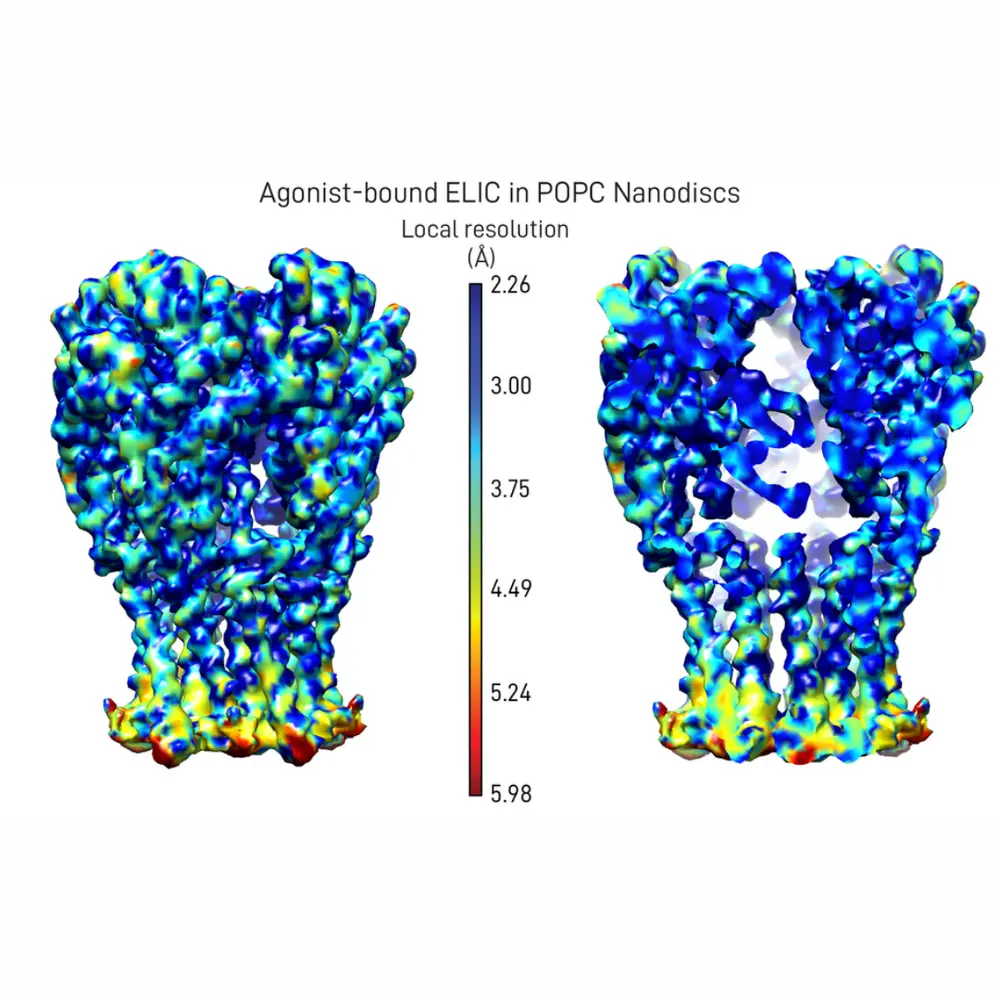
To find out how the new membrane affects the structure of ELIC, the authors determined two new structures using a structural-biology method called single-particle cryo-electron microscopy applied to this ion channel embedded in discs of lipid membrane. In ligand-gated ion channels (like ELIC), small molecules bind to the ligand-binding sites and favor a global conformational change that allows the protein to conduct ions. By comparing the two structures—one of which is ligand-bound, the other is not—the authors realized that the regions of ELIC near the binding sites are very different, while the transmembrane regions are similar. This suggests that the local membrane environment is capable of restricting the conformational changes of the transmembrane channel region, thus disrupting the coupling between the conformational changes undergone by the ligand-binding sites and the transmembrane pore. Using molecular dynamics simulations and free-energy calculations, they also found that the ligand-bound form of ELIC has a much higher binding affinity for the ligand than does the ligand-free form, as expected for a ligand that acts as an agonist.
“These ideas and methods are crucial in the context of the pharmaceutical industry”, Steven Wang added. The team was completed by Pramod Kumar (first author), Zhiyu Zhao, and Gisela D. Cymes, from the School of MCB, and Zhening Zhang, from the Simons Electron Microscopy Center, New York Structural Biology Center.


