The Freeman laboratory (Cell and Developmental Biology) delineates a molecular chaperone-dependent mechanism for selectively mobilizing gene loci through the nuclear actin matrix. Their findings were published in Developmental Cell.
A curious feature within all cells is the precise spatial organization of the genome. While early light microscopy work led to the concept of “chromosome territories”, electron microscopy as well as fluorescence in situ hybridization experiments validated the existence of structured chromatin fibers with non-random, higher-order packaging. Recent high-throughput molecular techniques have shown that eukaryotic genomes are partitioned into distinct compartments that are further divided into topologically associated domains, as well as smaller self-associating regions. Intriguingly, the exact packaging arrangement is dependent upon the health, age, and type of cell. Hence, genome organization promises to be key determinant for cell fate.
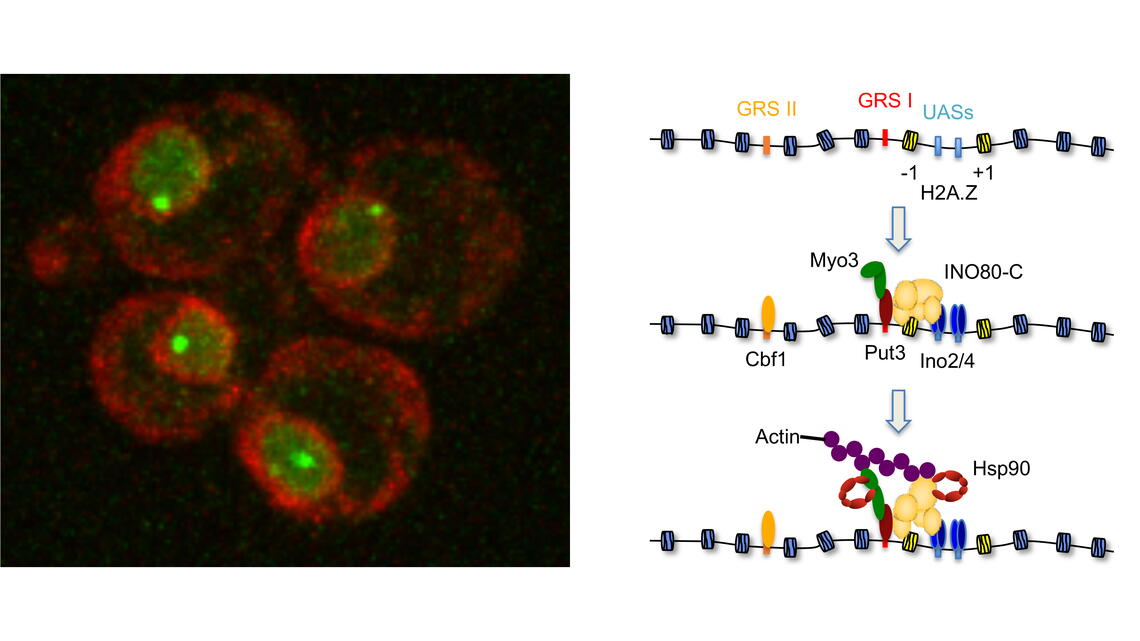
Despite the commonality of precise genome packaging, a mechanism for reorganizing chromatin in response to cellular demands had not been reported. The Freeman laboratory approached this problem by exploiting a simplified system in which a single gene locus is experimentally triggered to move from the inner nucleus to the nuclear periphery in response to physiological cues. Work led by Dr. Anqi Wang and Janhavi Kolhe delineated a molecular chaperone-dependent pathway for relocating activated gene loci. Their data support a model in which a two-authentication system mobilizes a gene promoter through a dynamic network of polymeric nuclear actin. Transcription factor-dependent nucleation of a myosin motor propels the gene locus through the actin matrix and fidelity of the actin association was ensured by ARP-containing chromatin remodelers. Motor activity of nuclear myosin was dependent on the Hsp90 chaperone. Hsp90 further contributed by biasing the remodeler-actin interaction towards nucleosomes with the non-canonical histone H2A.Z thereby focusing the pathway on select sites such as transcriptionally active genes. Together, the system provides a rapid and effective means to broadly yet selectively mobilize chromatin sites.
Read a review by Developmental Cell.