
Proximity labeling of cell structures followed by mass spectrometry has become an increasingly popular proteomics approach to identify what proteins localize to different cell structures. In practice, however, results are typically confusing, with long lists of hundreds of proteins identified, among which only a small fraction are bona fide components of the target cell structures.
An unappreciated problem in many of these studies has been the labeling radius assumed for the different proximity labeling methods. For example, methods employing peroxidases to generate a phenol-biotin (tyramide-biotin) free radical have generally assumed a staining radius of 20 nm. The Belmont laboratory at the University of Illinois, though, recently documented a staining radius of over 1000 nm. Joseph Dopie, a postdoctoral fellow in the Belmont laboratory, introduced a simple modification of the proximity labeling proteomics approach that implicitly takes into account this large staining radius to sort the list of identified proteins to identify the proteins most likely enriched in the target cell compartment.
The research was highlighted in the article, Tyramide signal amplification mass spectrometry (TSA-MS) ratio identifies nuclear speckle proteins, published in the Journal of Cell Biology this fall.
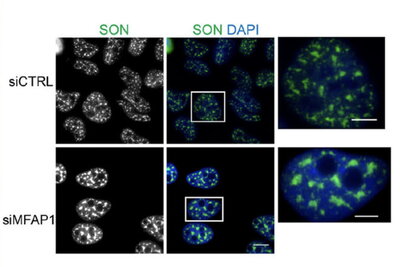
Using Tyramide Signal Amplification (TSA), in which horseradish peroxidase generates the free-radical proximity labeling, Dopie reexamined the proteomics of nuclear speckles. By comparing the ratio of protein abundance in nuclear speckles versus centromeres, he was able to distinguish proteins enriched in nuclear speckles versus in the nucleoplasm surrounding both nuclear speckles and centromeres. Of the top 100 sorted proteins, among thousands identified, ~70% had already been described in the literature or public data bases as nuclear speckle enriched. Using light microscopy, Dopie validated several additional proteins as being nuclear speckle enriched. This included MFAP1, a protein that enters reforming nuclei very soon after mitosis, may help nucleate the reformation of nuclear speckles after mitosis, and whose levels he showed modulate the size, stability, and dynamics of nuclear speckles.
Building on this data set, Dopie and others in the Belmont laboratory already have begun to identify proteins in structures surrounding nuclear speckles that may be involved in regulation of gene expression. Their preliminary results formed the foundation of a recently awarded grant proposal to identify new “transcriptional niches” within interphase nuclei.