University of Illinois scientists have identified the phosphorylation site and its consequence on the function of MOV10, an RNA helicase expressed in early brain development and required for embryo viability, illuminating this RNA binding proteins’ relation to microRNA pathways and its effect on protein expression.
Stephanie Ceman, a professor in the Department of Cell & Developmental Biology in the School of Molecular & Cellular Biology, and Aatiqa Nawaz, a PhD candidate in the Ceman Lab, have discovered that substitution of the serine at position 970 to mimic phosphorylation in MOV10 causes a conformational change that allows the protein AGO2 to access bound mRNAs, leading to their degradation.
MOV10 is a protein that is essential for gastrulation, a process key to early development of the fetus. When the MOV10 gene was knocked out in frog embryos in previous research by Ceman and colleagues, the embryo would shockingly extrude its contents and die before being able to fully develop, showing how important MOV10 was for embryonic development, Ceman said. This finding is particularly important because 95 percent of proteins are targets of microRNAs, and a disruption in microRNA pathways could greatly affect many proteins within human beings. Therefore, it is important to ask how MOV10 function is regulated.
“We collaborated with Dr. Jan Pohl’s group at the Center for Disease Control and Prevention to perform mass spectroscopy in order to identify the phosphorylation sites,” she said.
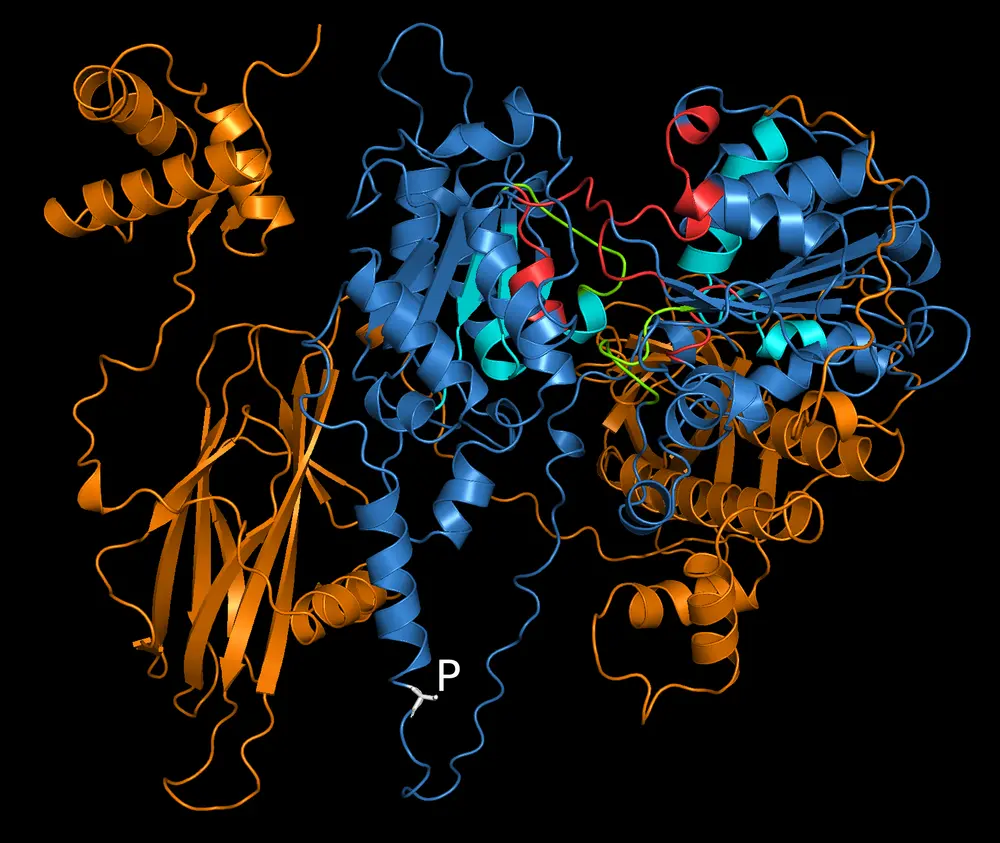
AlphaFold, the newly developed artificial intelligence system for protein structure prediction, was also integral to the research, and allowed the team to visualize MOV10 folding in relation to the phosphorylation site. The team discovered that the phosphorylation site was like “standing on the edge of a cliff of unstructured, unpredictable sequences,” Ceman said.
The team then found evidence that when phosphorylated, the conformation of the protein changes such that it opens for the AGO effector proteins access to the mRNA with consequent degradation. When not phosphorylated, the conformation of the protein is simply closed, allowing translation of bound mRNAs. In addition, phosphorylation inhibits the ability of MOV10 to unfold RNA secondary structures called G-quadruplexes.
Ceman, whose lab delves into the genetics of learning and memory via understanding the proteins required for those functions, has dedicated her research since her postdoctoral days to Fragile X syndrome.
“It revolves around a single gene that is involved in cognition, which makes it easier to study,” Ceman said. The protein that is absent in Fragile X syndrome is an RNA binding protein FMRP. In exploring FMRP function, Ceman’s group discovered that MOV10 interacted with FMRP. The end goal of this research is to be able to better understand the relationship between MOV10 and neuronal function, with real neuronal testing rather than through the neuroblastoma cell-line that they currently use, she said. The identification of S970 phosphorylation will help understand the role of MOV10 in neuronal function.
The paper, “Serine 970 of RNA helicase MOV10 is phosphorylated and controls unfolding activity and fate of mRNAs targeted for AGO2-mediated silencing,” was published in the Journal of Biological Chemistry. Research was funded by the Kiwanis Neuroscience Research Foundation and the National Science Foundation.

