Biochemistry graduate student Pei Wang and Associate Professor Raven Huang have discovered a new bacterial RNA repair complex. The structure of the 270-kDa RNA repair complex revealed that it is built like a shopping mall, and RNA repair can be achieved having the damaged RNA visiting four active sites with a minimum travelling distance. The findings are published in Nature Communications.
To fight for a limited resource of nutrition in the wildness, microbes release toxins to kill their neighbors to increase their share of nutrition. A majority of toxins are ribotoxins that cleave essential RNAs involved in protein translation, which are conserved in and required for all organisms to live. To counter the damage inflicted by ribotoxins, organisms employ protein enzymes to repair the damaged RNA for survival. Over the past several years, the Huang group has been engaged in discovery, biochemical and structural characterization of RNA repair systems. Significant research includes the discovery of a Pnkp–Hen1 RNA repair complex that carries out RNA repair with immunity. In the present study, published in the journal Nature Communications, Huang and coworkers described the discovery and characterization of a new bacterial RNA repair complex.
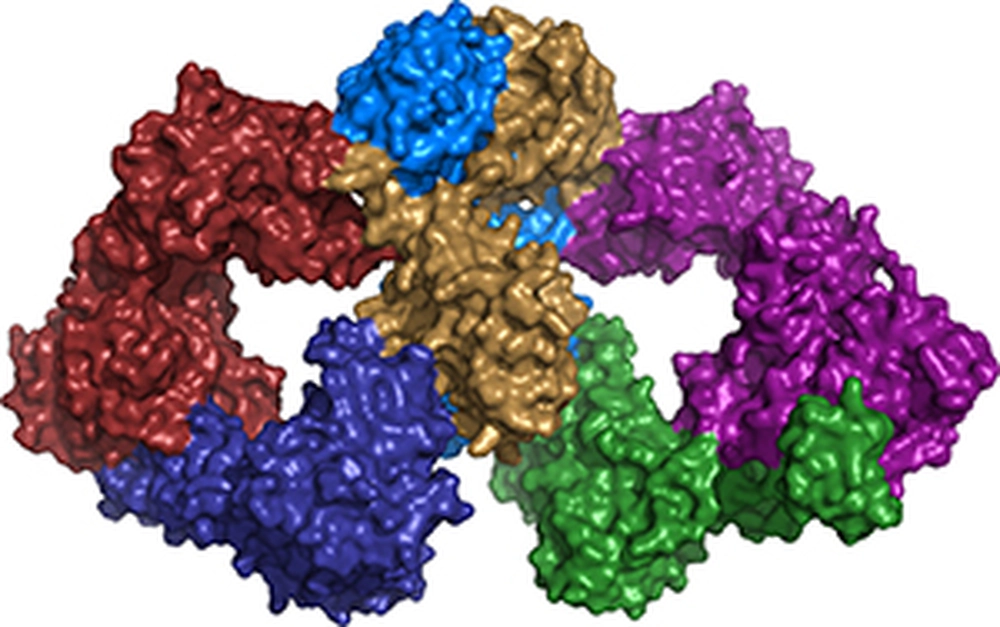
The approach involved bioinformatic analyses to reveal the presence of a new putative RNA repair system in certain bacteria. The genes encoding three proteins (named Pnkp1, Rnl, and Hen1) that constitute the putative new RNA repair system were then cloned into expression vectors. All three proteins were overexpressed in E. coli and purified to homogeneity. In vitro reconstitution using the purified recombinant proteins and a ribotoxin-cleaved RNA substrate demonstrated that RNA repair requires the presence of all three proteins. Furthermore, the three proteins were shown to form a heterohexamer in vitro. Huang and coworkers were able to crystallize and solve the structure of the Pnkp–Rnl–Hen1 heterohexamer. The structure revealed the molecular architecture of the heterohexamer as two ring structures of Pnkp1–Rnl–Hen1 heterotrimer fused at the Pnkp1 dimer interface. The four active sites required for RNA repair are located on the inner rim of each ring, reminiscent of architectures of shopping malls. This particular arrangement of the four active sites suggests that RNA repair might be carried out via a “one-stop shopping” mechanism.
Unlike the Pnkp–Hen1 RNA repair complex, which is present in many bacteria, the newly discovered Pnkp1–Rnl–Hen1 RNA repair complex is only found in ten bacterial species so far. Interestingly, the majority of the bacteria possessing the newly discovered RNA repair complex live in gingival plaques of human mouth. Huang hypothesized that the unique RNA repair carried out by the Pnkp1–Rnl–Hen1 complex in these bacteria might provide them with a heightened ability to survive. If this hypothesis proves to be correct, inhibiting RNA repair might provide a vehicle to reduce the population of these bacteria, which are linked to human dental and gum diseases. Development of small-molecule inhibitors of the new RNA repair complex is currently underway in Huang laboratory to test this possibility.