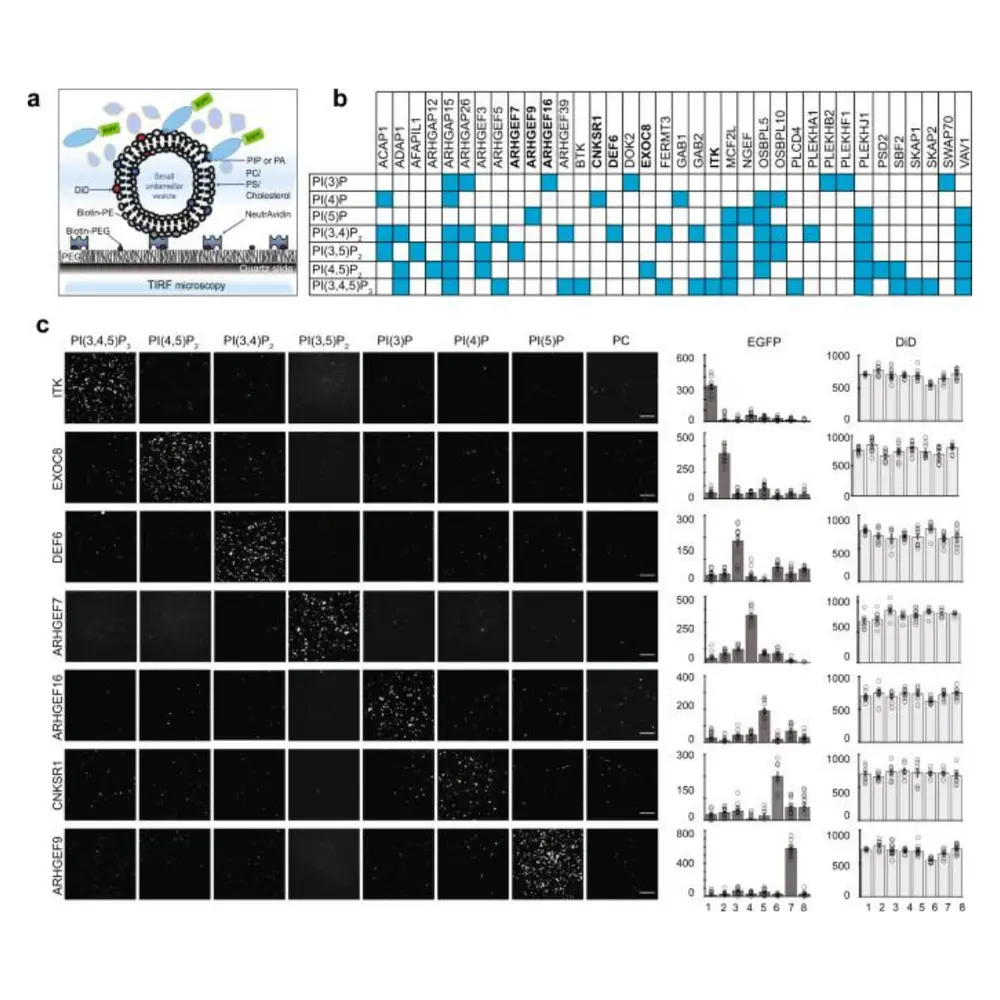
Phospholipid-protein interactions play an essential role in the regulation of many important cellular processes. The largest family of putative lipid-binding proteins contain the pleckstrin homology (PH) domain. Previous studies in the field estimate that approximately 10 percent of the PH protein family binds to phosphatidylinositol phosphate (PIP) with high specificity and affinity. However, new research from the University of Illinois School of Molecular & Cellular Biology contradicts that current view.
In their article, “Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins,” Jie Chen, professor of cell and developmental biology; Nilmani Singh, a recent PhD graduate; research specialist Adriana Reyes-Ordoñez and colleagues suggest that about 50 percent of PH domain-containing proteins interact with specific PIPs. The research was recently published in Nature Communications.

While PIPs only make up a small portion of the cell membrane, their role in cellular regulation is notable. Previously Chen and her team developed a novel single-molecule assay called Lipid-SiMPull to study protein-lipid interaction. Taekjip Ha, a professor of biophysics and biomedical engineering at Johns Hopkins University, was an important collaborator in that development. Many of the current methods that examine PH-PIP binding require protein purification and focus on the PH domain alone rather than the whole protein. Lipid-SiMPull offers a unique complement to the currently available methods, according to Chen. Her team took advantage of the SiMPull assay that relies upon Total Internal Reflection Fluorescence (TIRF) microscopy to visualize which proteins bound to the lipid vesicles of interest. The assay allows researchers to study full-length PH domain-containing proteins in whole-cell lysates without purification.
Because working with lipid vesicles is inherently challenging, Chen and her team encountered many technical obstacles during their study of protein-PIP interactions. However, after collecting data from over 5,000 assays for nearly 100 proteins of this family, they gained novel insights into their PIP binding characteristics. Using the assay data, they further pioneered a probabilistic sequence comparison algorithm, which can predict with high accuracy which PH domain sequences will bind to PIPs. After analyzing the amino acid sequences of 242 human PH domains, they determined that about half of them bind PIPs. These assay-based and assay-validated computational predictions have set Chen’s research apart and offer an interesting perspective on PH-PIP interactions.
Many of the lipid-binding proteins Chen’s assay and algorithm analyzed are involved in disease. For example, an obscure protein that binds PIP3 is overexpressed in ovarian cancer. Researchers speculate that this protein is a receiver for this lipid known to be associated with oncogenic activities. Further research on phospholipid-protein interactions could provide some insight into the mechanisms of these diseases. Additionally, due to the regulatory roles of PH-PIP binding, a greater understanding of their interactions could lead to drug development for such diseases.
Their groundbreaking research has challenged current views of PH-PIP interactions. In the future, Chen and members of her lab would like to implement machine learning to analyze PH domain sequences. Chen sees this project as the beginning of the further exploration of this lipid family. She hopes the scientific community is inspired by their discovery of so many PIP-binding PH domain-containing proteins and they pursue further research in the field.
This work was supported by grants from the National Institute of General Medical Sciences.

