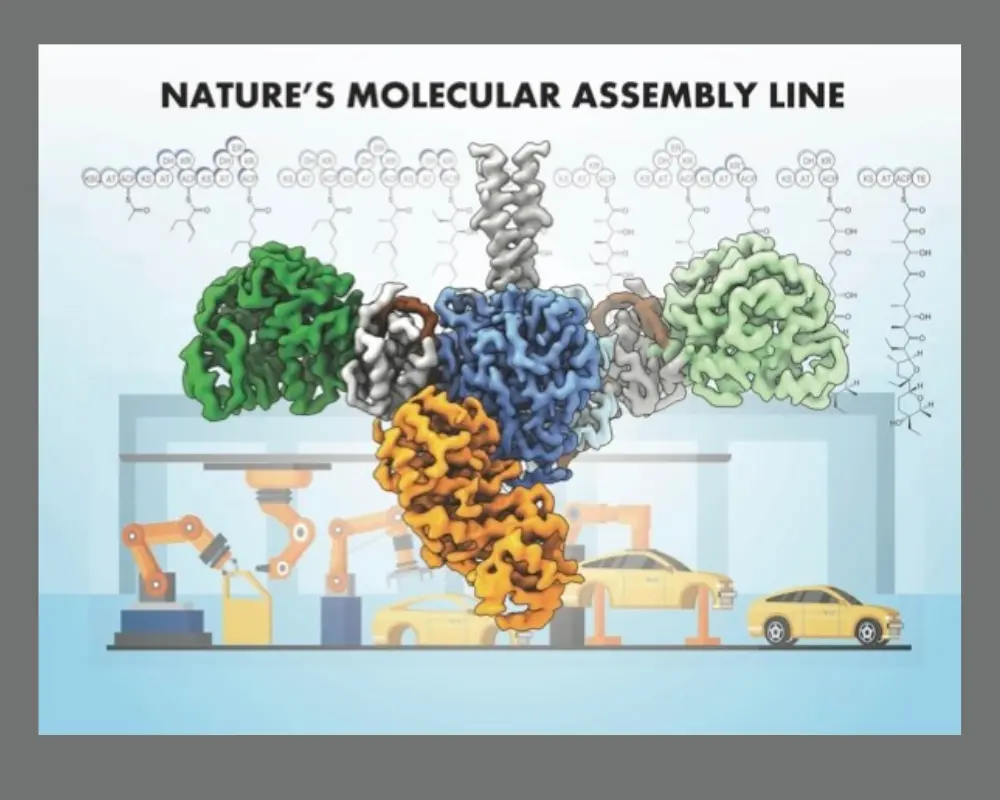
A new review article co-authored by Chu-Young Kim, Professor of Biochemistry, discusses the architecture of modular polyketide synthases (PKS), a type of enzyme found in various organisms that synthesize widely used clinical drugs like the antibiotic erythromycin and the immunosuppressant rapamycin. The article, which appears in Natural Product Reports, also features cover art submitted by the authors.
Kim recently spoke with the School of MCB Communications Office about the review, offering insight into its potential applications in drug discovery and development.
Why are researchers interested in understanding modular PKS architecture?
Most drugs are designed and synthesized by scientists. But antibiotics and anti-cancer drugs are often obtained by prospecting. Scientists go into fields or oceans hoping to discover a new cancer drug or a new antibiotic. That has worked very well for nearly 100 years, but now we are often rediscovering drug molecules that already exist. It’s a lot of time and effort to find out that what you’ve been after is something you already have.
The alternative approach is to do this in the lab. Modular PKSs are a type of enzyme that make polyketide antibiotics and anti-cancer drugs, among others. We want to know what these enzymes look like structurally so that we can engineer them. And if you successfully engineer PKSs, you can trick bacteria into producing polyketide drugs which do not exist in nature or which do exist but have not been discovered yet. This way, you don’t have to go into nature anymore; you can do everything in the lab. It’s definitely more convenient, and it saves time and money. But engineering PKSs hasn’t been easy; it doesn’t always work as well as we’d hope. I think the main problem is that we have been engineering PKSs without knowing what they look like. It’s like trying to build a house without a blueprint. The recent high-resolution PKS structures provide blueprints that scientists can use to create engineered enzymes.
How do modular PKSs construct polyketide natural products?
They do it in an assembly line fashion, similar to how cars are built in auto plants. In the system we are studying, seven PKS enzymes make up the drug assembly line. The first enzyme builds the first portion of the drug, and then this partial drug molecule is transferred to the second PKS enzyme in the assembly line to be elongated. After all seven, you have the end product.
Your review emphasizes that polyketides are difficult to synthesize using chemical methods. Why is that?
They are very large. Size is a problem because it takes longer and is more expensive to produce. If you were to synthesize these drugs chemically, you might end up paying $100,000 for a 10-day course of antibiotics; they wouldn’t be accessible for everybody. Polyketide drugs also contain many chiral centers, which makes their chemical synthesis especially challenging. But enzymes are very good at dealing with chiral centers, which is why PKSs are very useful.
What were some benefits and challenges you encountered while using deep learning to map polyketide architecture?
The PKS structures were originally determined using X-ray crystallography or cryo-electron microscopy. Afterwards, we used AlphaFold, an AI tool developed by Google scientists, to predict the PKS structures and compared the predicted structures to the experimentally determined structures. AlphaFold is easy to use. You simply input the amino acid sequence of your protein, and AlphaFold will output the predicted three-dimensional structure of that protein in just a few hours. We were curious to know how well this works for PKSs. The results were mixed; AlphaFold correctly predicted the general shape of two of the three PKSs, but it got the details wrong for all three.
With the current technology available, it seems like these AI tools are useful if you just want a rough idea of what your protein might look like. But if you need highly accurate and detailed structural information for drug development or protein engineering, it’s not quite there yet. To be fair, AlphaFold is very good at predicting structures of small and simple proteins. But the modular PKSs are large and complex, so it’s a challenge for the program.
Tell us more about the X-ray crystallography approach.
I like X-ray crystallography because it routinely generates high-resolution structures. And for certain tasks, you need that high-resolution structural information. But the disadvantage is that the protein has to be crystallized first. You can’t simply have the protein in solution, or it won’t work. So, you have to coax the protein into becoming a crystal, which can be quite challenging for large or flexible proteins.
Were you surprised by your results?
Yes, modular PKSs are a lot more structurally complex than we envisioned. The protein we worked on, Lsd14, has a total of eight active sites. Most enzymes have just one active site. In the Lsd14 structure, we see a large number of inter-domain hydrogen bonds and salt bridges that presumably facilitate communication between the various active sites. To properly engineer PKSs, we’ll have to consider these interactions and complexities.
What is still unknown about the architecture of these PKSs?
So far, scientists have only resolved the structure of small PKS proteins, those with eight or less active sites. But we also need to know what the larger PKSs look like. We are now investigating a PKS protein that has 20 active sites, and another with 32 active sites. These studies will push the limits of X-ray crystallography and cryo-electron microscopy.
What do you want others to take away from this?
Many of the PKS structures in the review paper were determined using cryo-electron microscopy. The main advantage of this technique is that you do not need to crystallize your protein. The School of MCB now has a state-of-the-art cryo-electron microscope that anyone can access. I would encourage my colleagues and students to utilize this exciting resource in their research.
Main image features cover art provided by Chu-Young Kim and Saket R. Bagde.
